Mapping Environmental Justice in the Muhheakunnuk Valley
We’ll explore environmental justice in Dutchess County, within the Muhheakunnuk or Hudson Valley region.
Purpose
You will:
Load tabular and spatial data in R.
Plot data using ggplot geoms.
Use a spatial join to attach Toxic Release Inventory (TRI) facilities to neighborhoods (tracts).
-
Create a simple exposure indicator. Compute:
- TRI facilities per tract
- TRI per square mile
Explore associations between TRI facility locations and neighborhood indicators such as asthma prevalence, unemployment, and the percentage of children in a neighborhood.
Create choropleth maps of Dutchess County, NY using
ggplot.
Install packages
- You only need to do this step once.
Load the required packages
First, let’s use the read_sf function from the sf package to read in spatial data…
Step 1: Load spatial data
We will work with two spatial datasets:
dutchess_indicators: Census tracts with demographic and socioeconomic indicators.tri_facilities: TRI facilities (point locations).
# County variables (e.g., median household income)
#dutchess_indicators <- read_sf("data/dutchess_indicators.geojson")
# Point locations of TRI facilities for the Hudson Valley
#tri_facilities <- read_sf("data/toxic_release_inventory_epa.shp")
# Make sure that both layers are projected on the earths surface
# correctly: they need to have the same projection
tri_facilities <- st_transform(tri_facilities,
st_crs(dutchess_indicators))Explore exposure to pollutants from toxic release facilities
Explore and compare the location of the U.S. Environmental Protection Agency’s list of facility location producing or working with toxic chemicals.
-
tri_facilitiescontains one row for each Toxic Release Inventory (TRI) facility in the Hudson Valley. We will work with those in Dutchess County, NY.
Dutchess County, NY Federal Data
For this activity we’ll use the Census Tract geography, which are roughly neighborhood-sized areal units.
Each observation represents a Census Tract “neighborhood” and contains various social, economic, and built environment attributes for that neighborhood.
Our data
dutchess_indicators includes:
Join identifier for each Census Tract:
join_IDTotal population:
tot_popArea in square miles:
area_sqmilesMedian Household Income:
median_hh_incomeAsthma prevalence among adults:
asthma_rate
Each row in tri_facilities represents the registration of a site that emits toxic material into the land, air, or water.
Display the names of the variables in the dutchess_indicators dataset.
Step 2: Get to know the data
Explore the variables
Create ggplot charts to summarize the data.
First, let’s explore the numerical distribution of some of our numerical variables…
Numerical distribution
Histogram
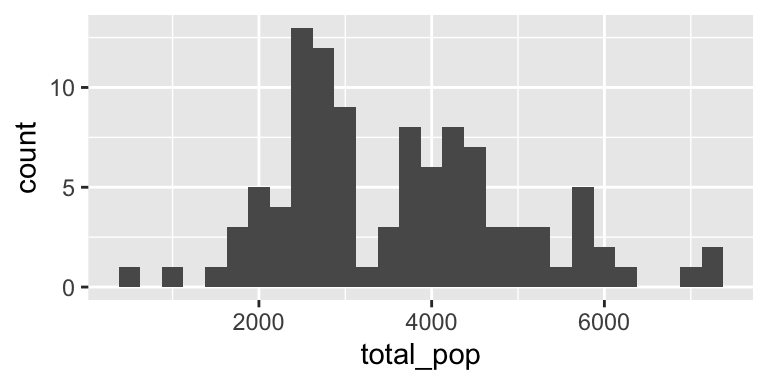
Density plot
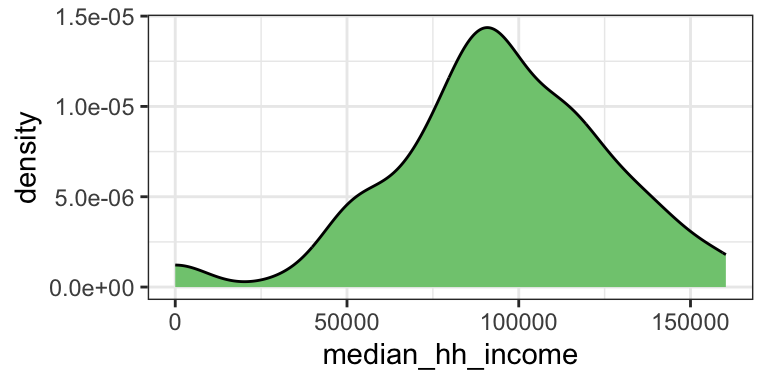
Boxplot
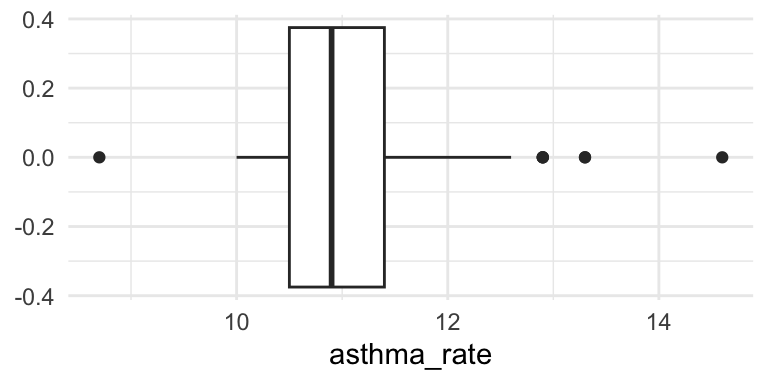
Maps!
Map the point locations of TRI facilities
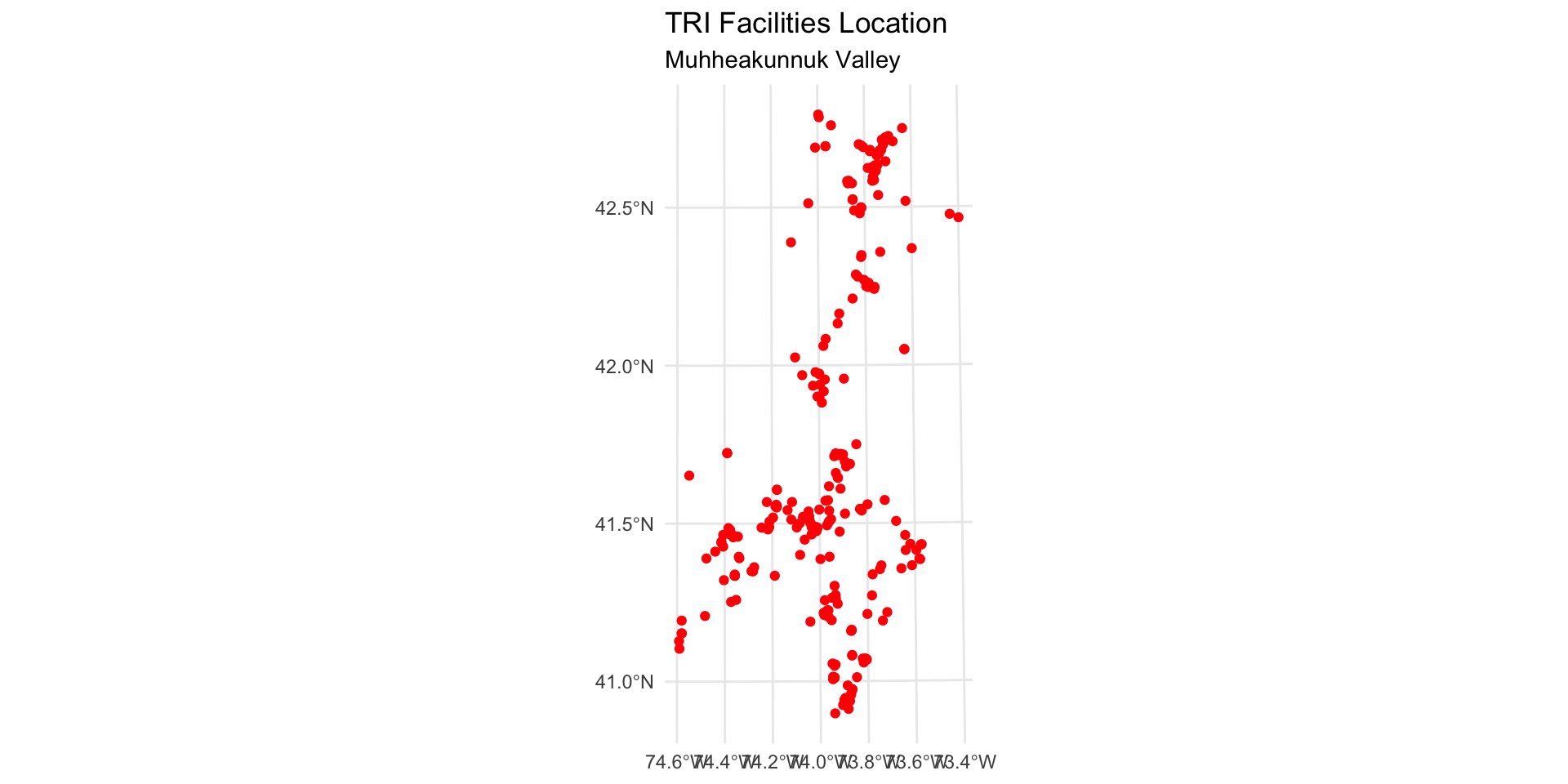
Map the point locations of TRI facilities
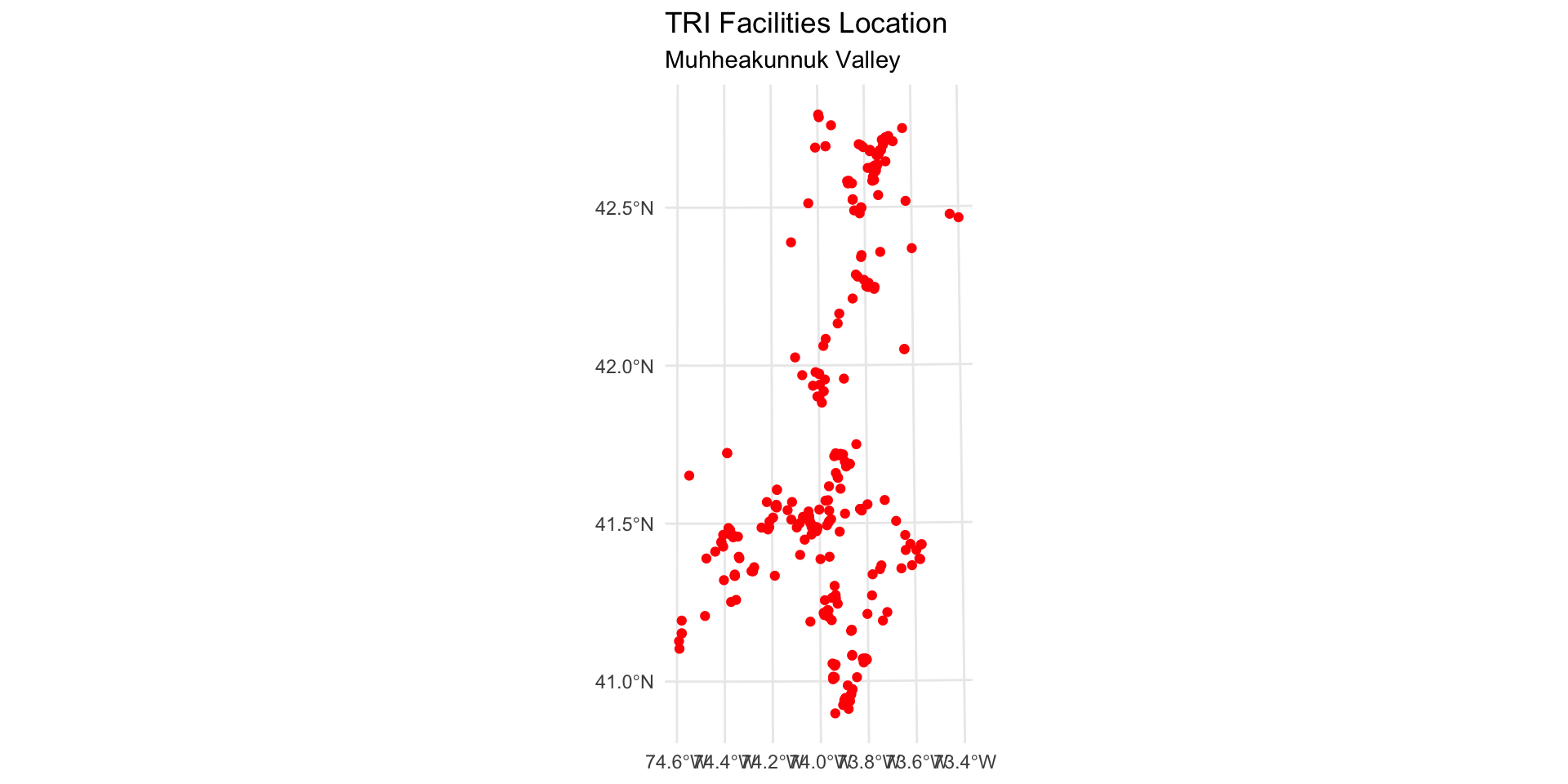
Map the point locations of TRI sites
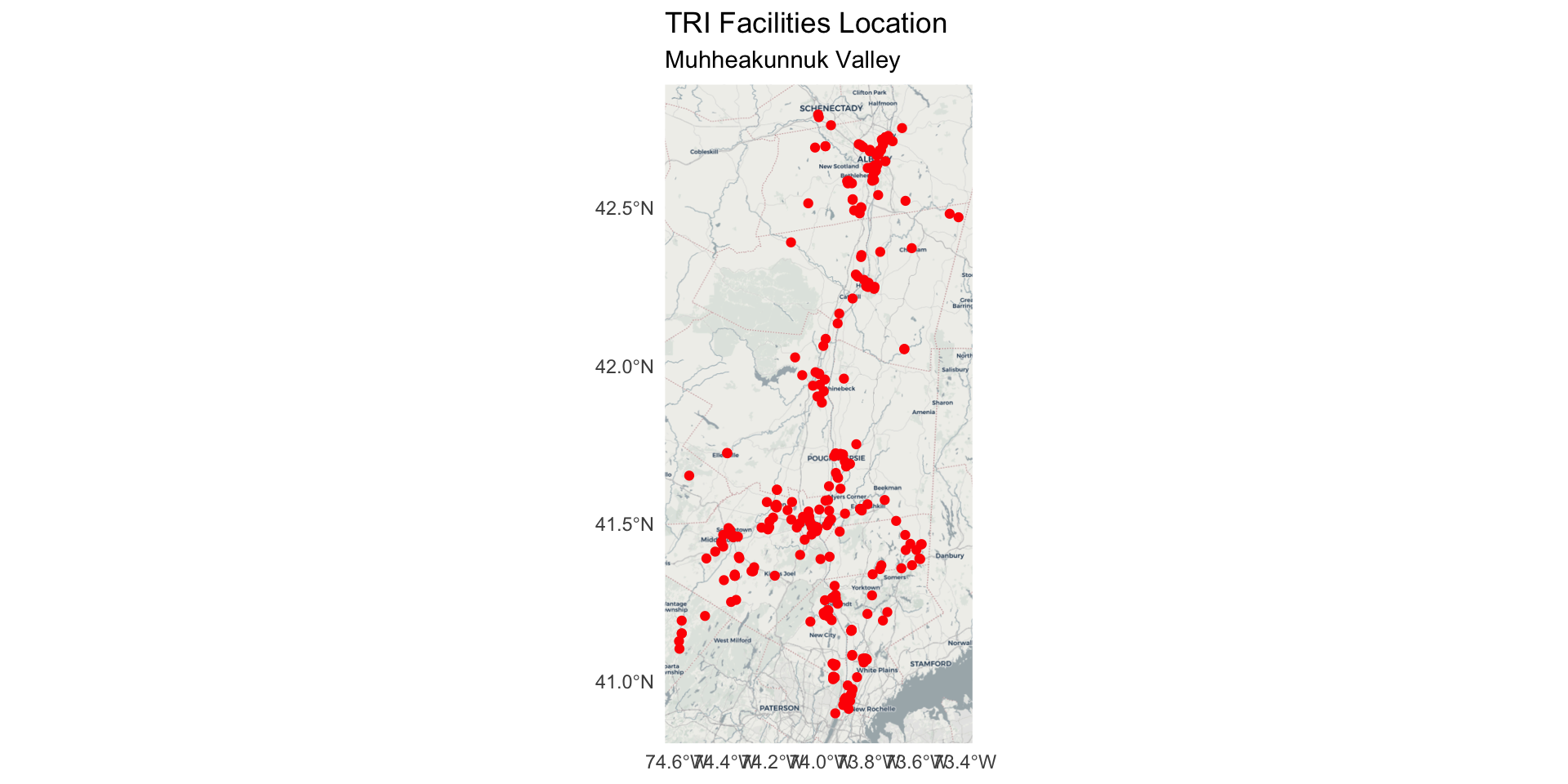
Map the point locations of TRI sites
This time, with a basemap.
Install and load the
ggspatiallibrary.Set the
zoom =argument to display a clear, readable base map underneath the geom_sf() point layers.
Spatial join
st_join() adds the columns from one spatial dataset to another based on how their shapes overlap or relate in space.
Use ?st_join to learn more.
Examples of spatial join relationships
One point falls inside a polygon
Two polygons touch each other
A line crosses a boundary
One feature is the nearest to another
Spatial join
It uses intersects, meaning if shapes touch or overlap, they get joined.
It does not require a matching column
It does not change the shapes
Imagine placing transparent paper maps on top of each other. Where features line up, st_join() copies the information from one map onto the other.
Spatial join
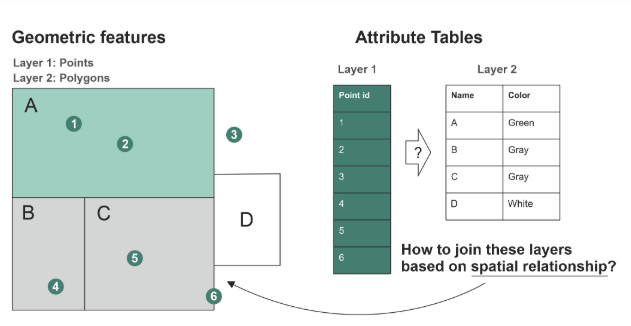
st_within and st_intersects argument inside st_join (Hint: use ?function_name to learn more.)
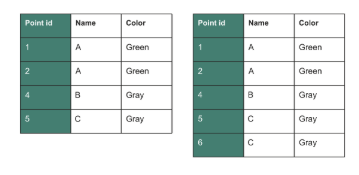
Summary of a spatial join
st_join()is a join by location, not by column.It adds attributes from one spatial layer to another.
It uses spatial relationships like intersects, within, contains, or nearest.
-
Used for the task of transferring attributes across overlapping layers:
Assigning census data to point locations (e.g., TRI sites)
Matching point data attributes to neighborhoods
Use the same coordinate system
In order to conduct any spatial operations like st_join both layers need to be projected onto the earth surface in the same way.
- We do this using a coordinate reference system or
crs
k12_points <- st_as_sf(k12_df, coords = c("x", "y"),
crs = 4326) %>% # Always load new data using the default crs 4326
st_transform(3626) # transform the coordinate reference system to be the same as dutchess_indicators and dutchess_tri
# Alternative approach
k12_points <- st_as_sf(k12_df, coords = c("x", "y"),
crs = 4326) %>%
st_transform(st_crs(dutchess_indicators))Step 3: Spatial join
Purpose: We want to know which neighborhoods contain TRI facilities. So, we attach each tract join_ID to the TRI facilities observations.
It tells which neighborhood (aka census tract) each TRI observation is within.
We use
st_join()with ast_withinpredicate to attach tract attributes to each TRI point.
Next, we use the result of the st_join to create a table that tells us how many TRI facilities are within each neighborhood.
We us the unique
join_IDto create a frequency table of TRI per tract.Then, we normalize by creating a variable that holds TRI facility density: TRI per square mile.
Step 4: Counting TRI facilities per tract
Now we aggregate to the tract level: how many TRI facilities are located in each tract?
What function do we need to get the number of observations per neighborhood (tract)?
Step 4: Counting TRI facilities per tract
Now we aggregate to the tract level: how many TRI facilities are located in each tract?
Step 5: Join and compute TRI facilities per square mile
Join the TRI per neighborhood counts back with the full dataset using a
left_joinby the unique join key in each data set:join_IDCreate a new varialble
tri_per_sqmilewith TRI facilities per square mile.Store the result in a new sf object
dutchess_tri
Summary statistics
Running the summary on a data frame object gives you the min, max, 1st quartile, median, mean, 3rd quartile, and max for each numerical variable.
total_pop area_sqmiles median_hh_income percent_children
Min. : 536 Min. : 0.1256 Min. : 0 Min. : 0.00
1st Qu.:2616 1st Qu.: 2.6370 1st Qu.: 78770 1st Qu.:15.20
Median :3502 Median : 5.5814 Median : 93278 Median :18.00
Mean :3609 Mean :11.0726 Mean : 94374 Mean :17.73
3rd Qu.:4522 3rd Qu.:14.3142 3rd Qu.:114663 3rd Qu.:21.50
Max. :7137 Max. :56.9210 Max. :160294 Max. :28.20
percent_white percent_black percent_latine unemployment_rate
Min. :11.20 Min. : 0.40 Min. : 1.70 Min. : 0.000
1st Qu.:64.40 1st Qu.: 2.70 1st Qu.: 7.90 1st Qu.: 2.300
Median :73.40 Median : 5.20 Median :11.50 Median : 4.200
Mean :68.98 Mean :10.28 Mean :13.02 Mean : 4.773
3rd Qu.:79.00 3rd Qu.:10.40 3rd Qu.:16.90 3rd Qu.: 6.850
Max. :89.50 Max. :60.80 Max. :30.90 Max. :16.100
asthma_rate low_food_access prox_traffic_pollution tri_n
Min. : 8.70 Min. : 0 Min. : 0.0 Min. :0.0000
1st Qu.:10.50 1st Qu.:2055 1st Qu.: 23.2 1st Qu.:0.0000
Median :10.90 Median :2964 Median : 79.8 Median :0.0000
Mean :11.09 Mean :3034 Mean : 183.9 Mean :0.4369
3rd Qu.:11.40 3rd Qu.:4074 3rd Qu.: 200.0 3rd Qu.:0.0000
Max. :14.60 Max. :6245 Max. :1719.9 Max. :6.0000
tri_per_sqmile geometry
Min. :0.0000 POLYGON :103
1st Qu.:0.0000 epsg:3626 : 0
Median :0.0000 +proj=tmer...: 0
Mean :0.1665
3rd Qu.:0.0000
Max. :2.2206 Step 6: Map TRI facilities per square mile
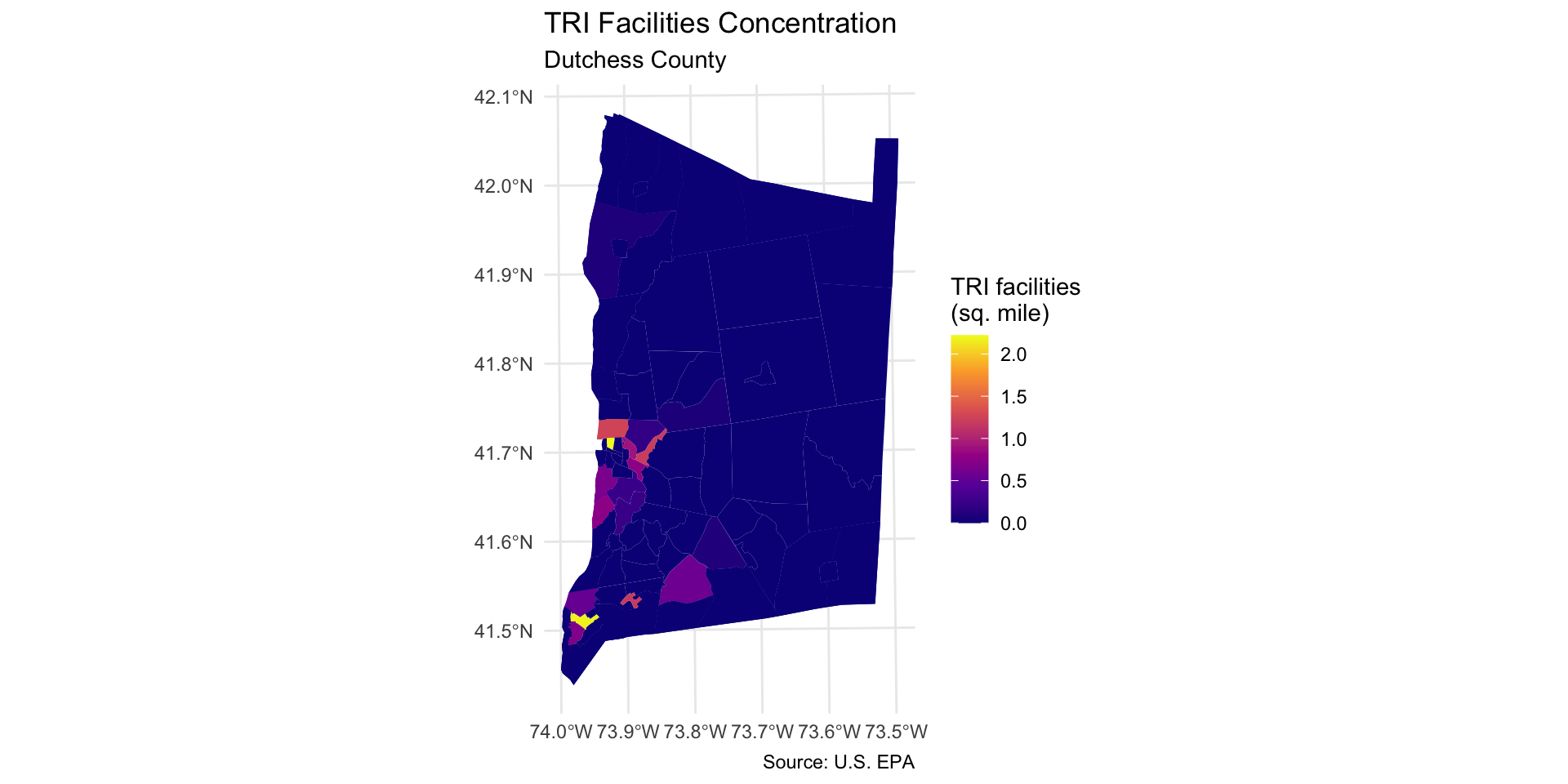
Step 6: Map TRI facilities per square mile
Map the count of TRI facilities per square mile across each neighborhood using geom_sf:
- Use
caption = "Source: U.S. EPA"to add a source note to the bottom of the map extent.
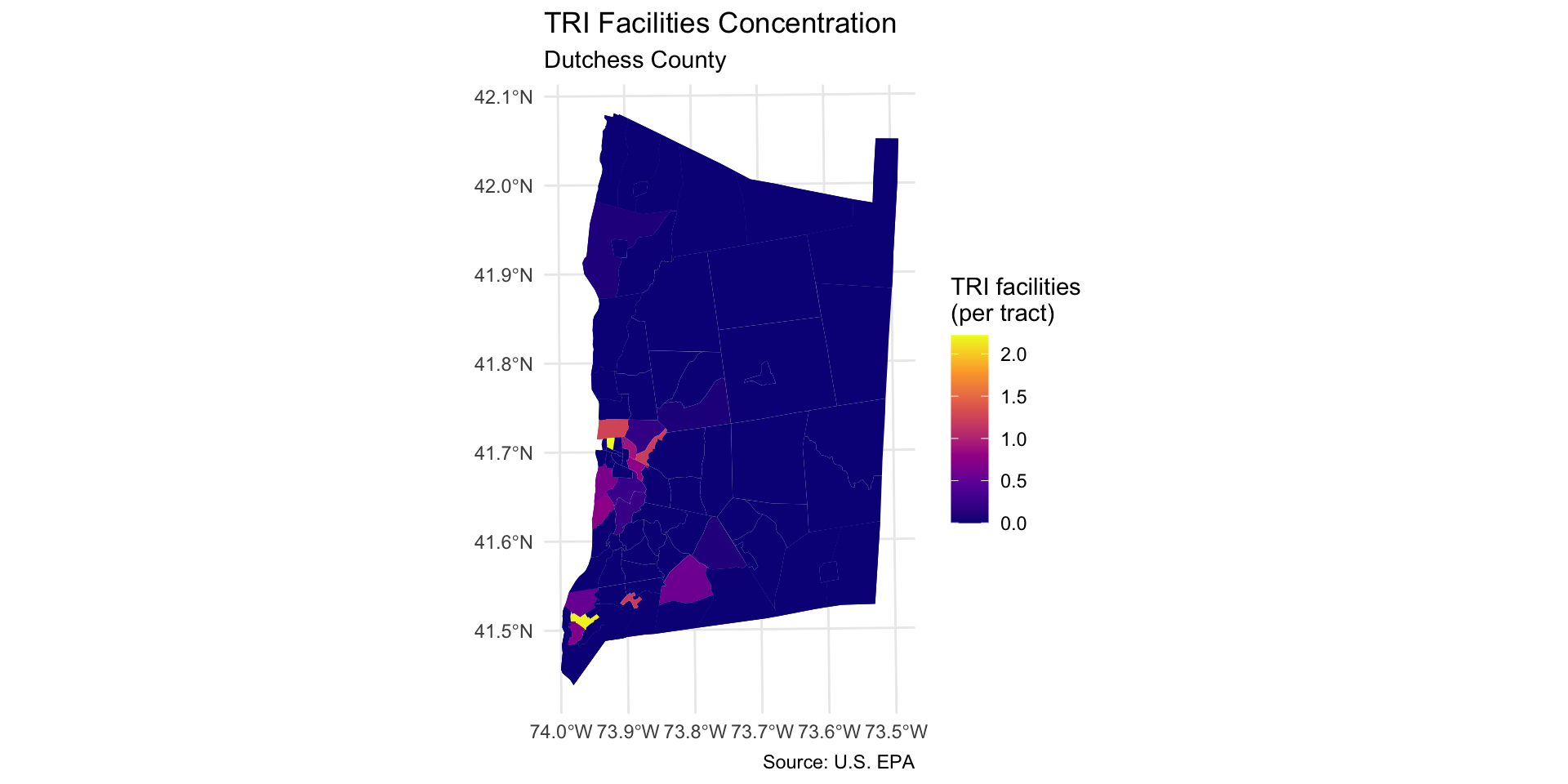
Step 7: Map other indicators of interest
Select another two or three variables from the dutchess_tri data frame to map. Change the fill = argument in the geom_sf() function to display the new indicator.
You will also need to change the fill legend title and title of the map.
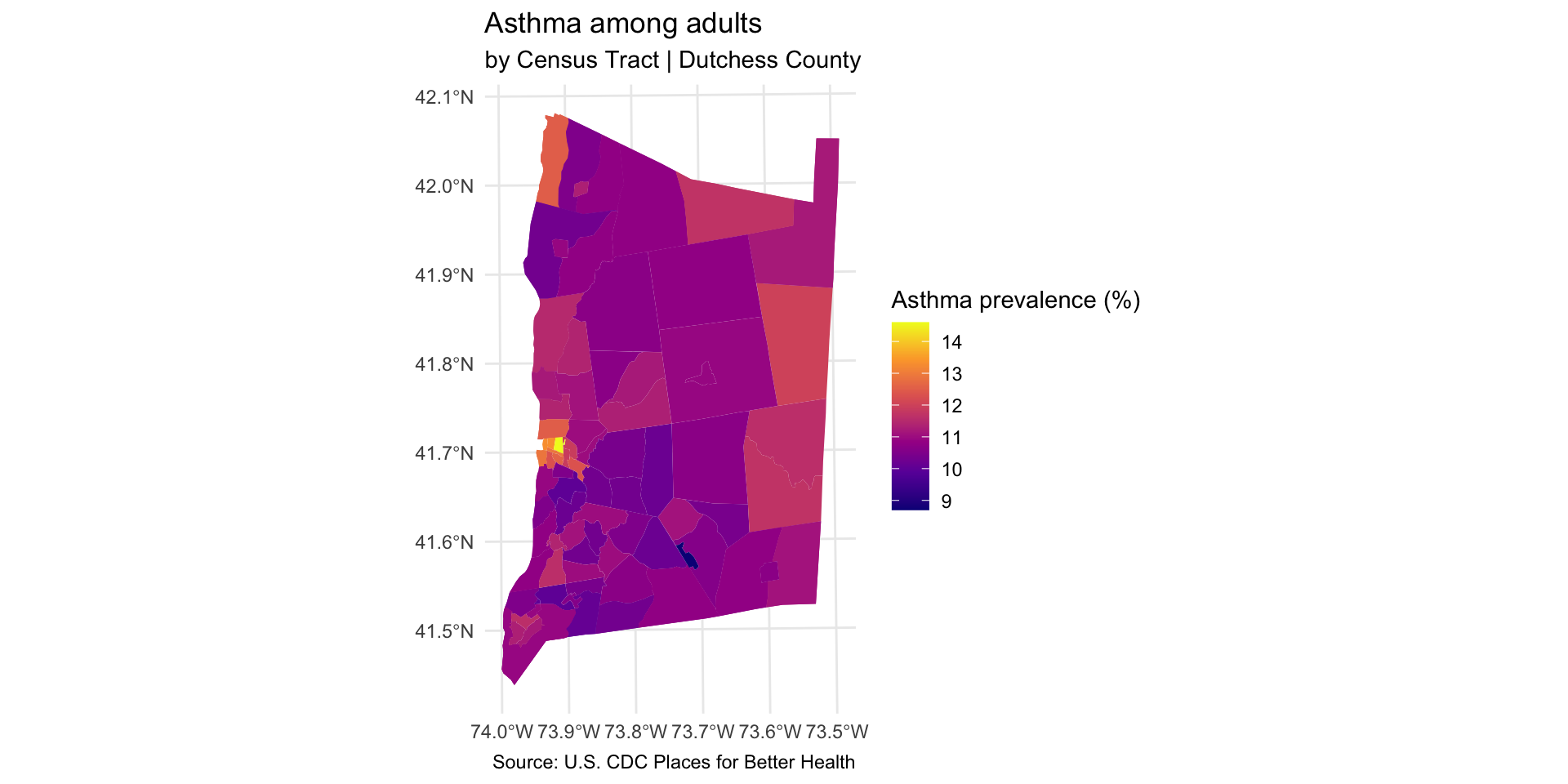
Step 8: Explore associations with neighborhood indicators
We can now explore how the location of TRI facilities is associated with neighborhood-level indicators.
Scatter plot to explore correlation
Compare TRI facilities tri_per_sqmile with the prevalence of asthma among the population: asthma_rate
Are neighborhoods with a higher concentration of TRI facilities near adults correlated with higher rates of asthma?
- Use
geom_smooth(method="lm") to add a linear regression line to your plot.
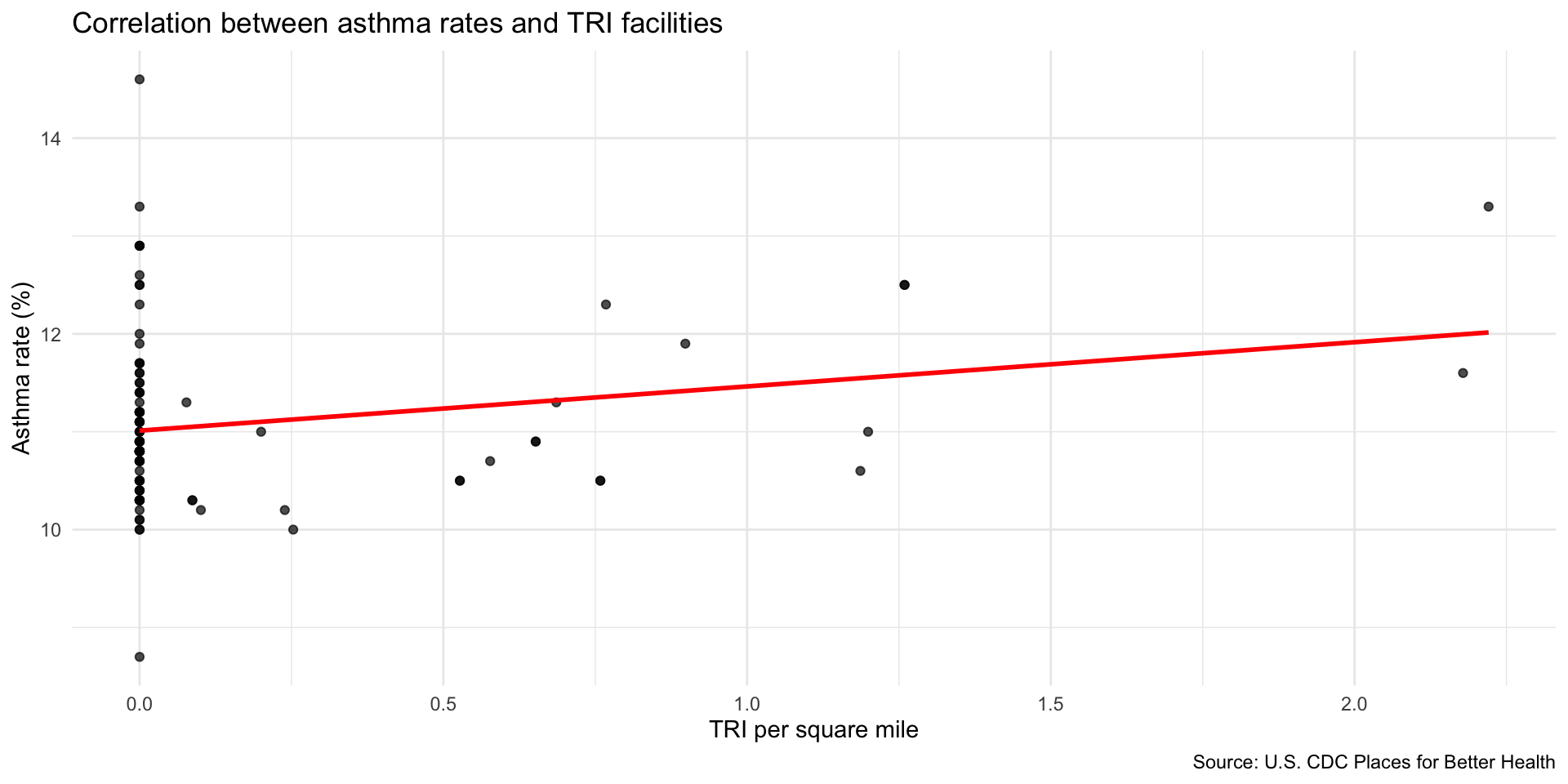
Use geom_jitter() to better observe the clustering of observations
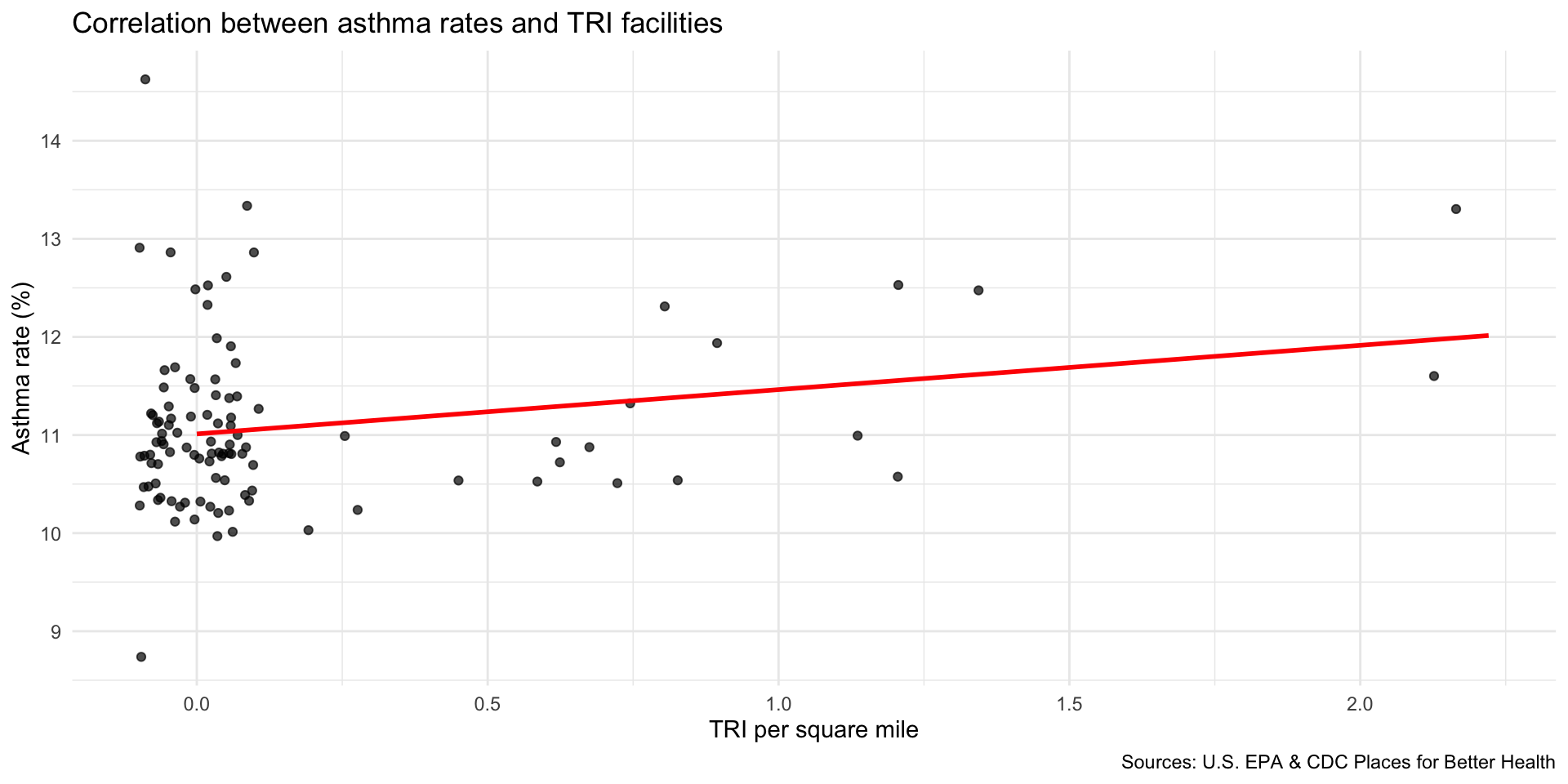
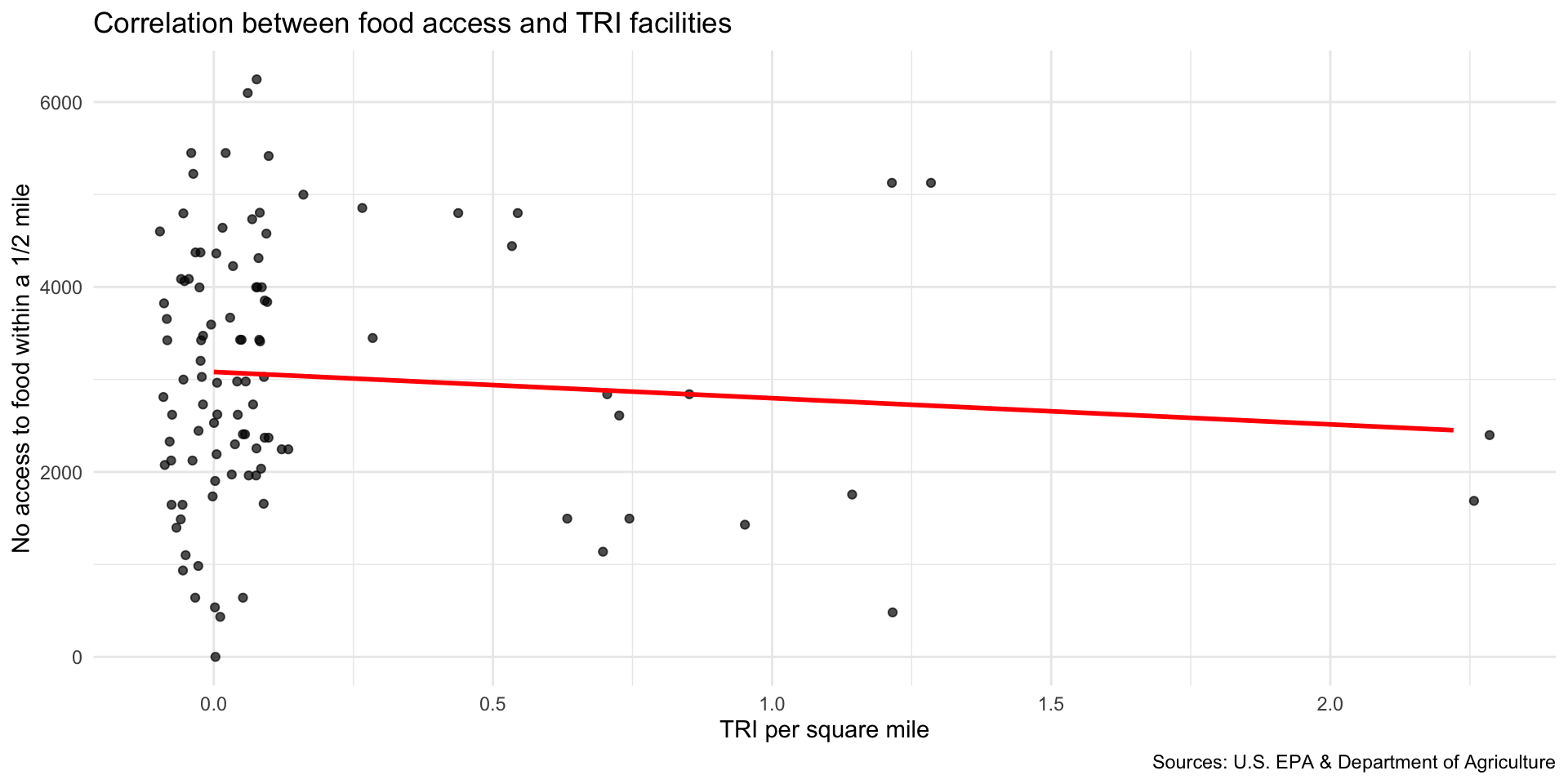
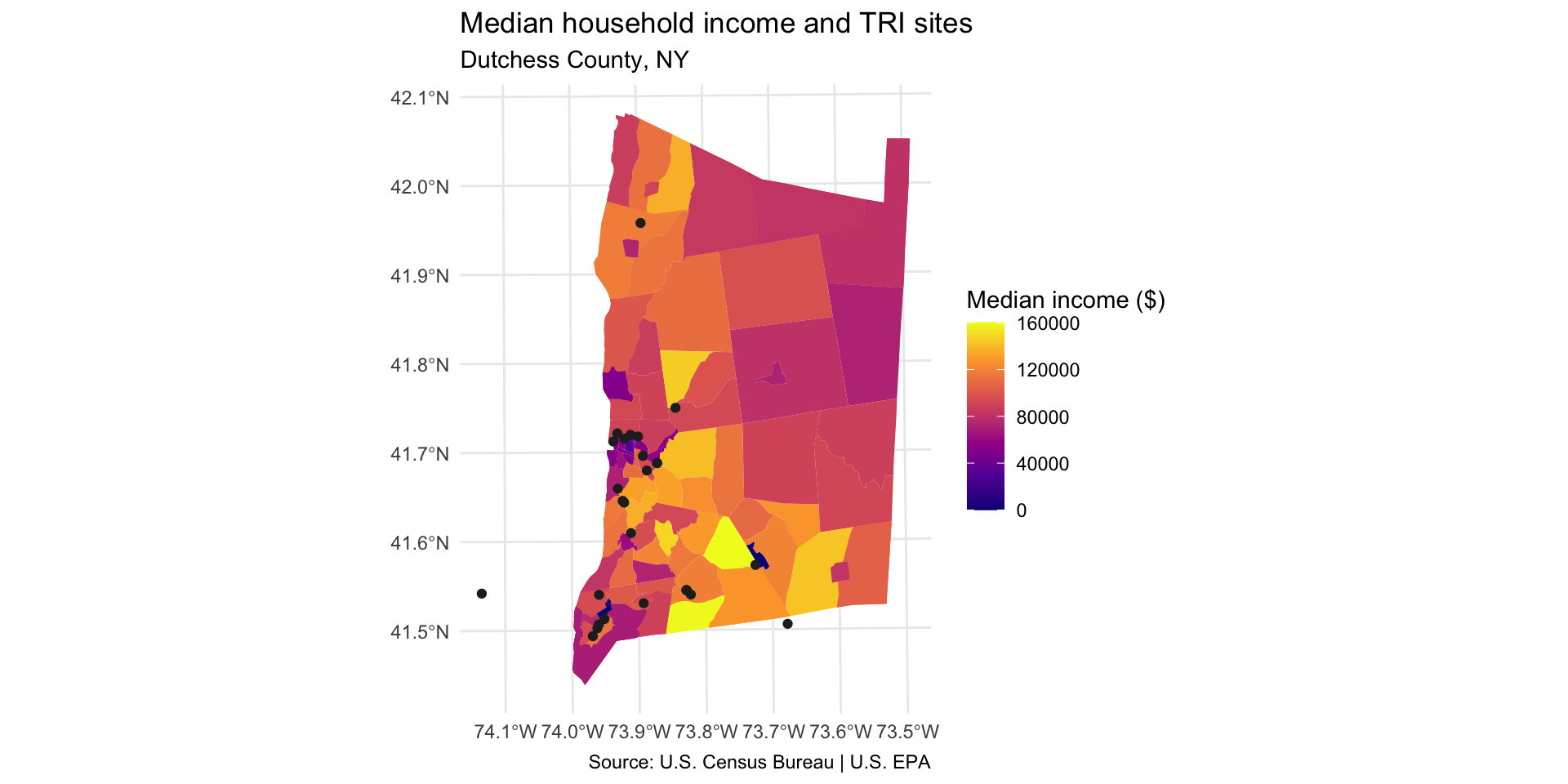
Step 9: Map TRI facility points with median household income
Compare TRI facility locations with the median household income in the neighborhood median_hh_income.
Step 9: Map TRI facility points with median household income
-
First, create a TRI layer that only includes facilities within Dutchess County.
Use the
COUNTY_NAMvariable to identify sites in Dutchess County.Use
distinctto identify how the Dutchess County entry is formatted in this dataset.NOTE: there is at least one TRI facility that appears outside of Dutchess County due to errors in the EPA database.
Create a TRI layer that only includes facilities within Dutchess County
COUNTY_NAM
1 ORANGE
2 ULSTER
3 COLUMBIA
4 GREENE
5 DUTCHESS
6 ORANGE COUNTY
7 COLUMBIA COUNTY
8 ULSTER COUNTY
9 SULLIVAN COUNTY
10 GREENE COUNTY
11 DUTCHESS COUNTY
12 PUTNAM
13 WESTCHESTER
14 ALBANY
15 ROCKLAND
16 BERGEN
17 RENSSELAER
18 SCHENECTADY
19 SUSSEX
20 ALBANY COUNTY
21 SUSSEX COUNTYMake an interactive map with the tmap package
Install and load the tmap package.
Note: you could use this interactive to identify the point(s) outside of Dutchess County (on the west of the estuary), then filter them out for the final map. For example, use `filter(REGISTRY_I != 110070690536)
Step 9: Map TRI facility points with median household income
In order to show two layers on the same ggplot map, you will need to use the
data =argument inside yourgeom_sf. For example:geom_sf(data = sf_dataset).Are neighborhoods with a higher concentration of TRI facilities more likely to have higher or lower median income?
Note: you will see that one TRI facility location showing up on the west side of estuary has an incorrect entry in the
COUNTY_NAMvariable. Ideally, we would remove this observation, but we can keep it in there for now.
dutchess_tri %>%
ggplot() +
geom_sf(aes(fill = median_hh_income), color = NA) +
scale_fill_viridis_c(option="C", name="Median income ($)") +
geom_sf(data = tri_dutchess) +
labs(title="Median household income and TRI sites",
subtitle="Dutchess County, NY",
caption = "Source: U.S. Census Bureau | U.S. EPA"
) +
theme_minimal()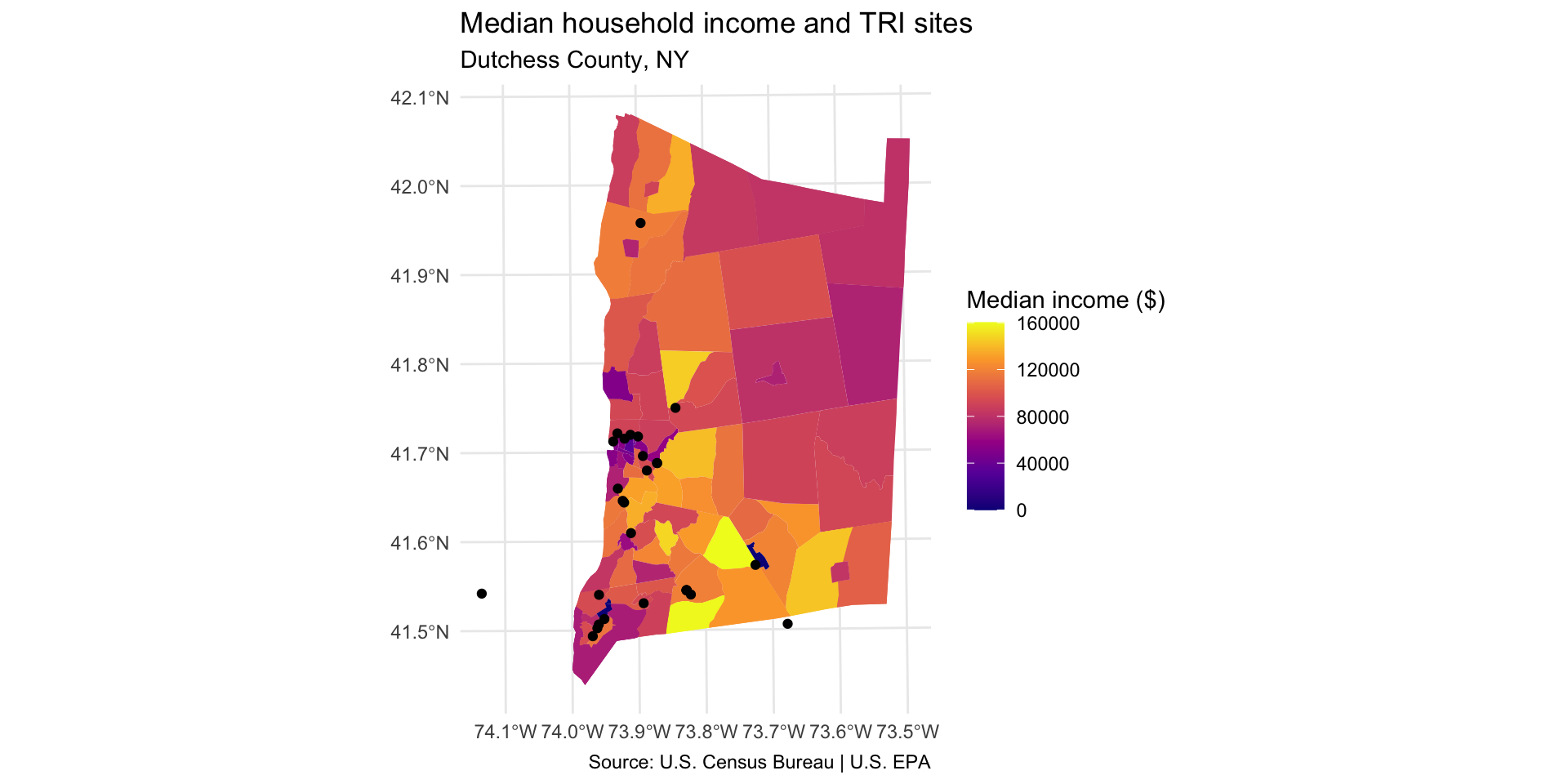
Add legend item for TRI facility points
-
Add the following to generate a legend item for the point layer:
aes(color = "TRI facilities")mapping function generates a legend itemSet color aesthetic in
scale_color_manual()
dutchess_tri %>%
ggplot() +
geom_sf(aes(fill = median_hh_income), color = NA) +
scale_fill_viridis_c(option="C", name="Median income ($)") +
geom_sf(
data = tri_dutchess,
aes(color = "TRI facilities"), # mapped aesthetic makes legend item appear
size = 1.5
) +
scale_color_manual(
name = NULL, # no legend title
values = c("TRI facilities" = "#252525")
) +
labs(title="Median household income and TRI sites",
subtitle="Dutchess County, NY",
caption = "Source: U.S. Census Bureau | U.S. EPA"
) +
theme_minimal()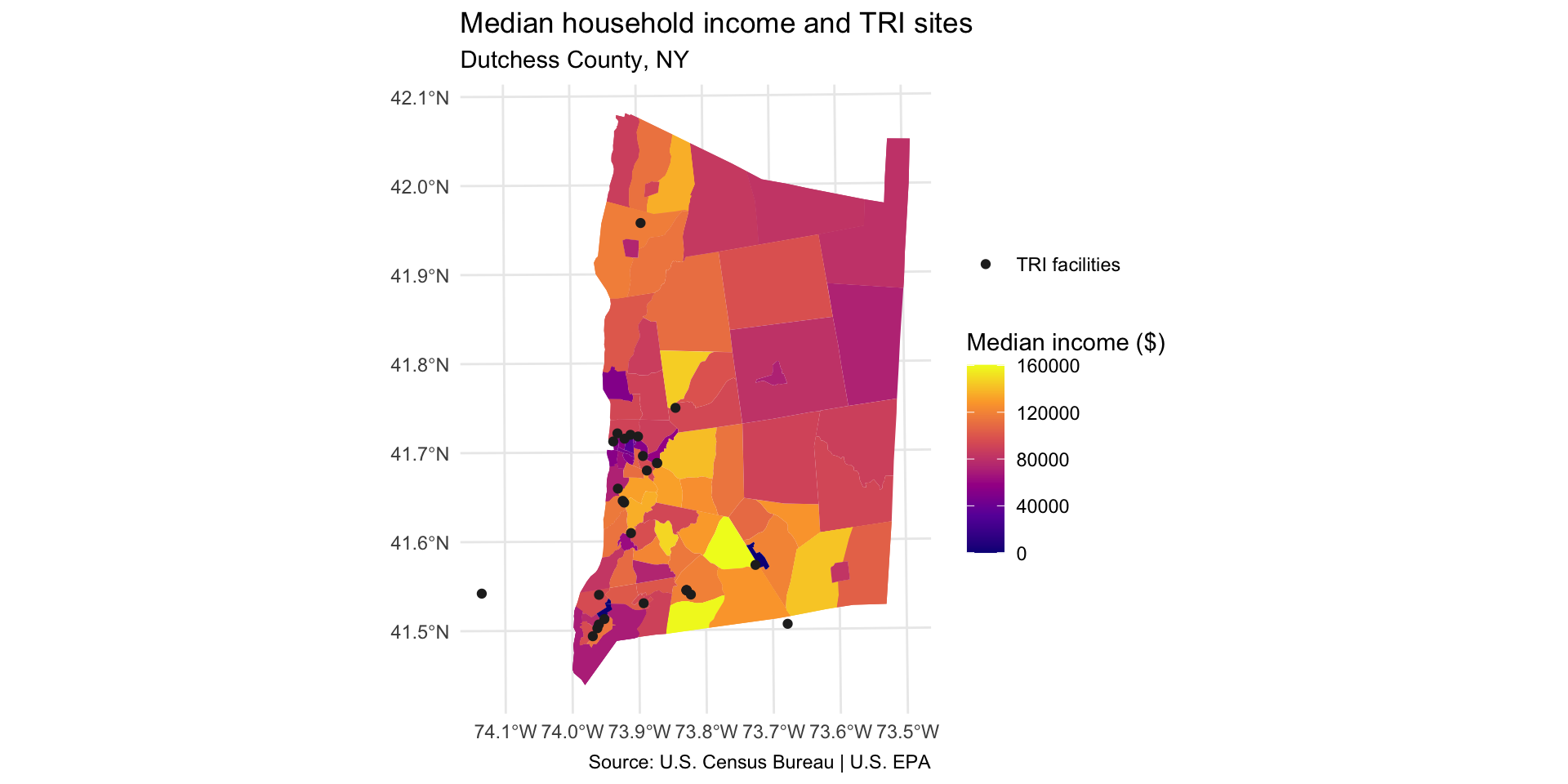
Discussion
Identify and describe which parts of the county have higher TRI density. Use cardinal directions (north, south, east, west).
Describe one pattern in terms of: income, asthma, or an indicator you choose.
Why do you think we observe these relationships? Come up with one hypothesis that explains the pattern you observe.
Share with the class
What pattern did you see?
What might explain it?