More aesthetics and facets
Intro to Data Analytics
ggplot aesthetics
Reminders: ggplot2 aesthetics
- Aesthetic characteristics can be mapped to variables
- color
- shape
- size
- alpha (transparency)
Reminders: ggplot2 aesthetics
- Aesthetics as mapping
- Determines size, color, etc. based on variables in the data
- Goes inside aes( )
- Aesthetics as setting
- Determines size, color, etc. based on fixed values, e.g, don’t depend on the values of variables.
- Goes inside geom( )
Data: Palmer Penguins
Measurements for penguin species, island in Palmer Archipelago, size (flipper length, body mass, bill dimensions), and sex.
Rows: 344
Columns: 8
$ species <fct> Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Adel…
$ island <fct> Torgersen, Torgersen, Torgersen, Torgersen, Torgerse…
$ bill_length_mm <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.3, 38.9, 39.2, 34.1, …
$ bill_depth_mm <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.6, 17.8, 19.6, 18.1, …
$ flipper_length_mm <int> 181, 186, 195, NA, 193, 190, 181, 195, 193, 190, 186…
$ body_mass_g <int> 3750, 3800, 3250, NA, 3450, 3650, 3625, 4675, 3475, …
$ sex <fct> male, female, female, NA, female, male, female, male…
$ year <int> 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007…Plot
Code
Coding out loud
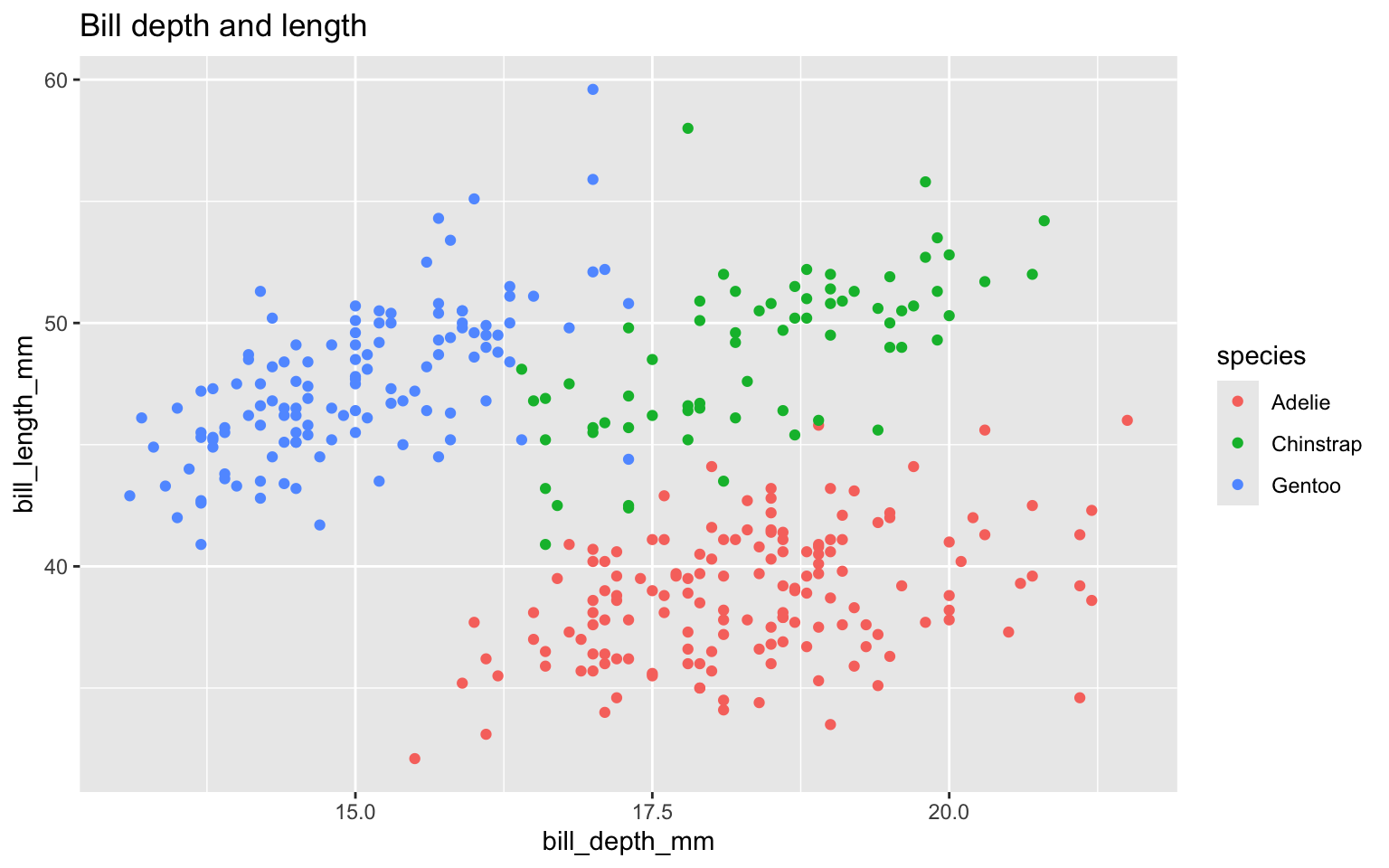
Start with the
penguinsdata frame
Start with the
penguinsdata frame, map bill depth to the x-axis
Start with the
penguinsdata frame, map bill depth to the x-axis, bill length to the y-axis.
Add scatter plot
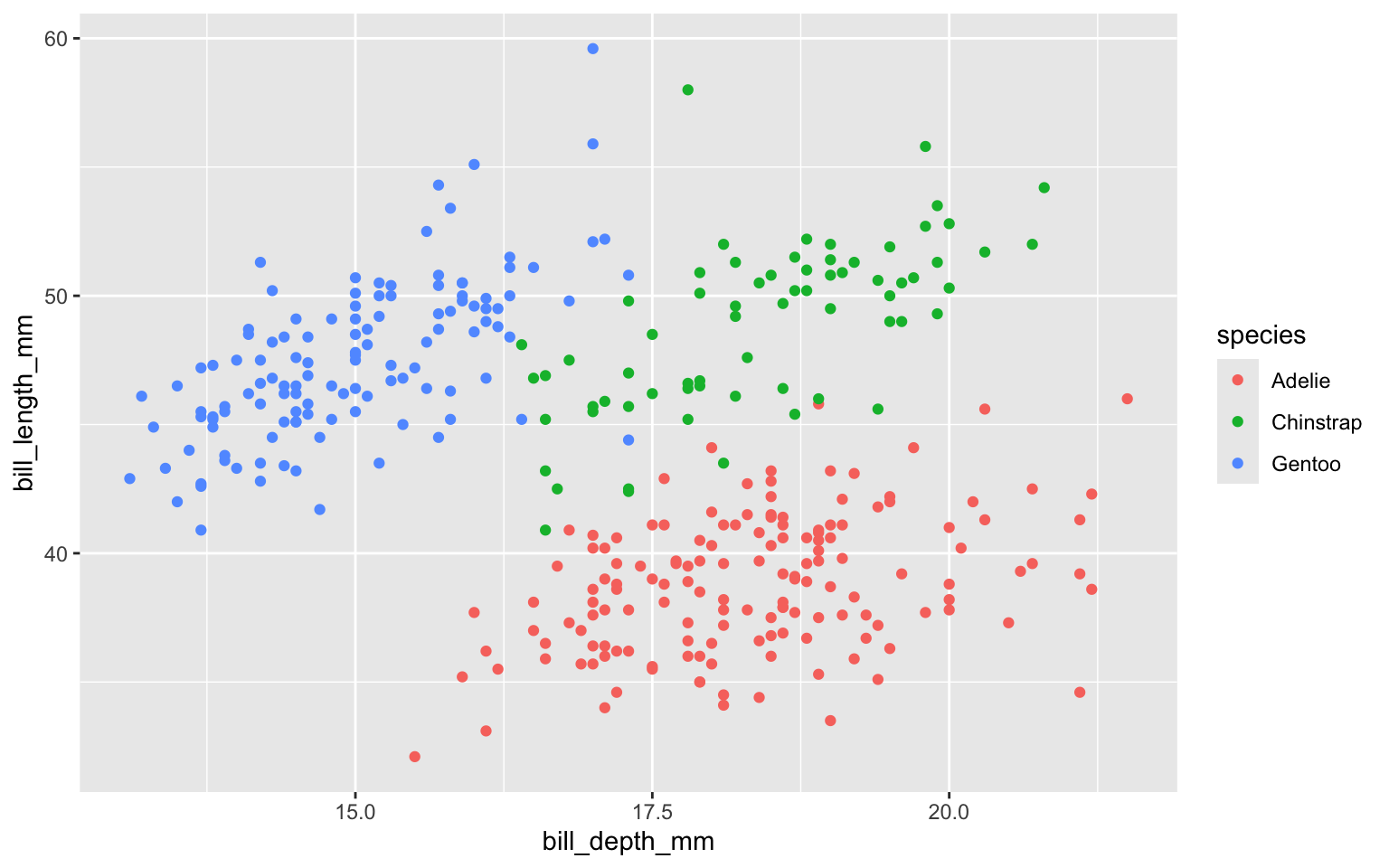
Color based on species (map species to the color of each point).
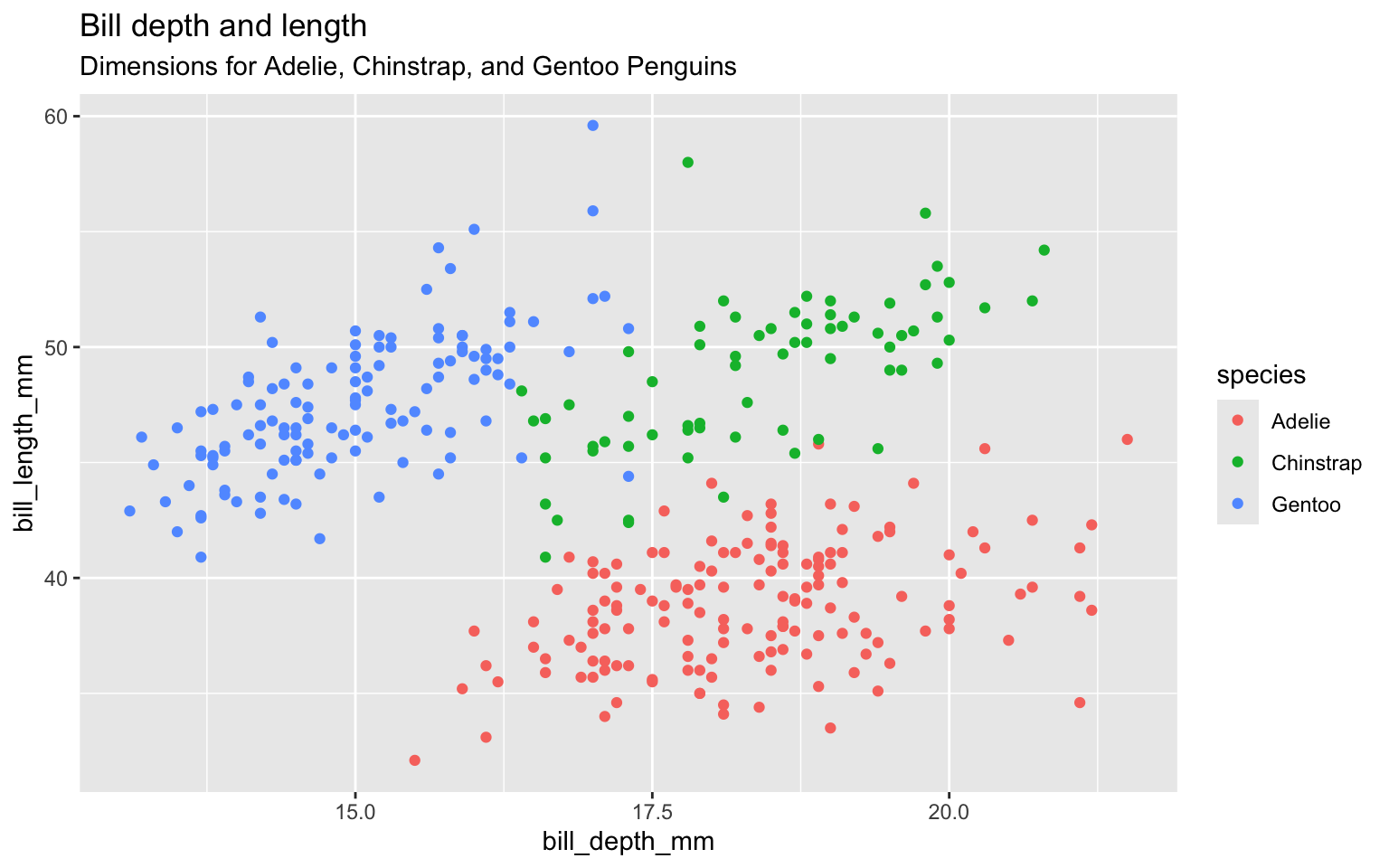
Title the plot
Add the subtitle
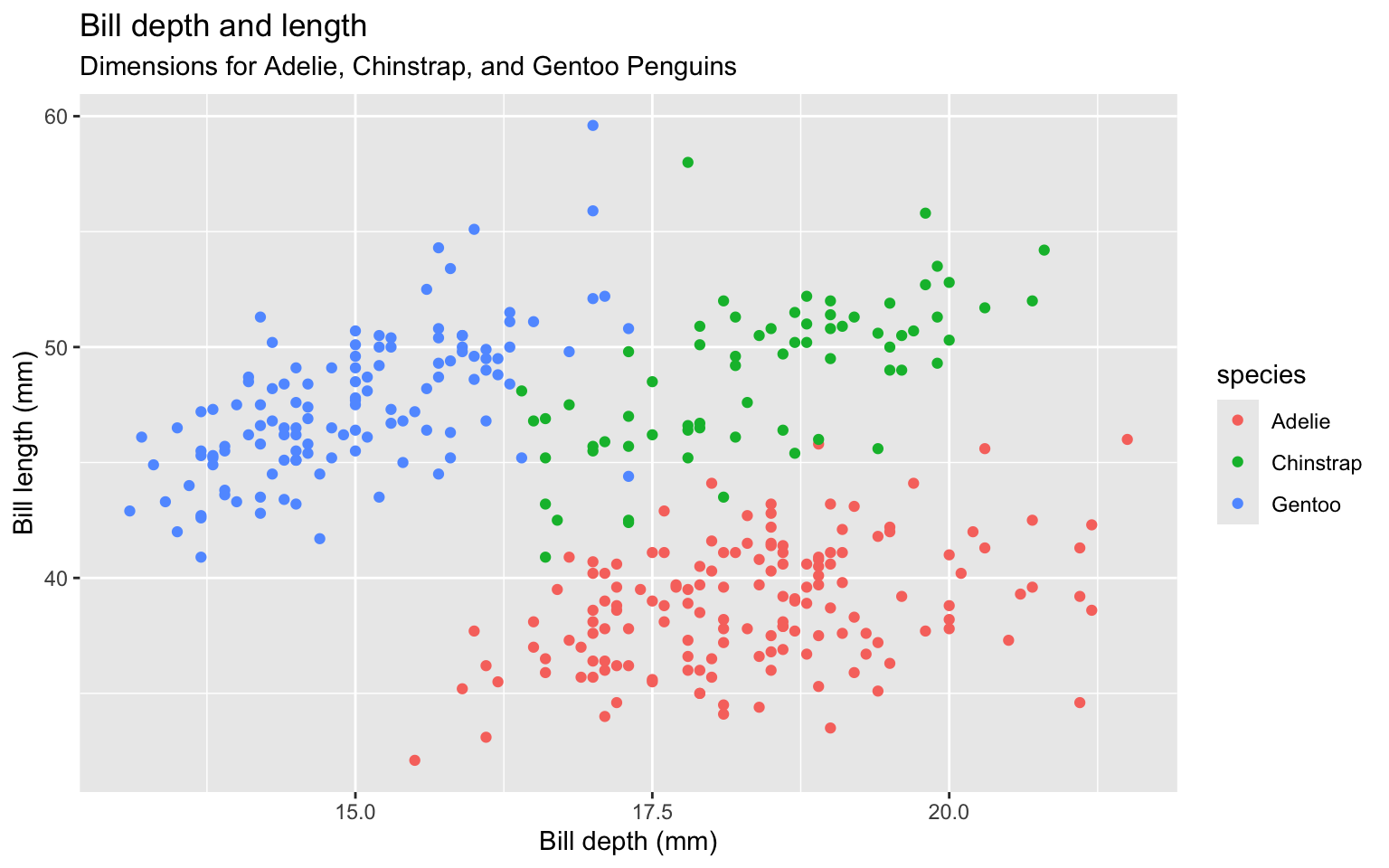
Clarify the graphic by labeling the x and y axes
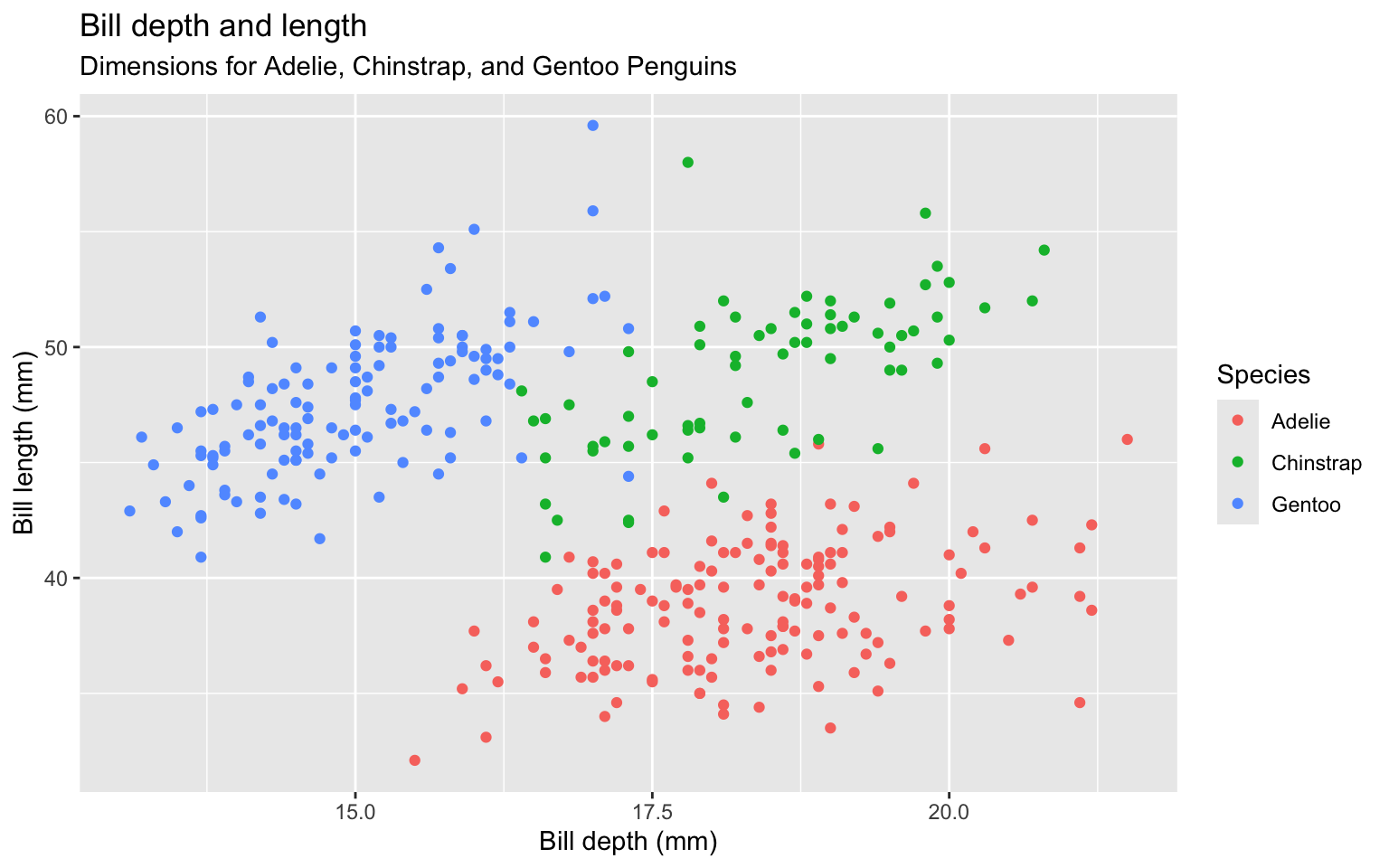
Label legend “Species” instead of using column name
Add a caption for the data source.
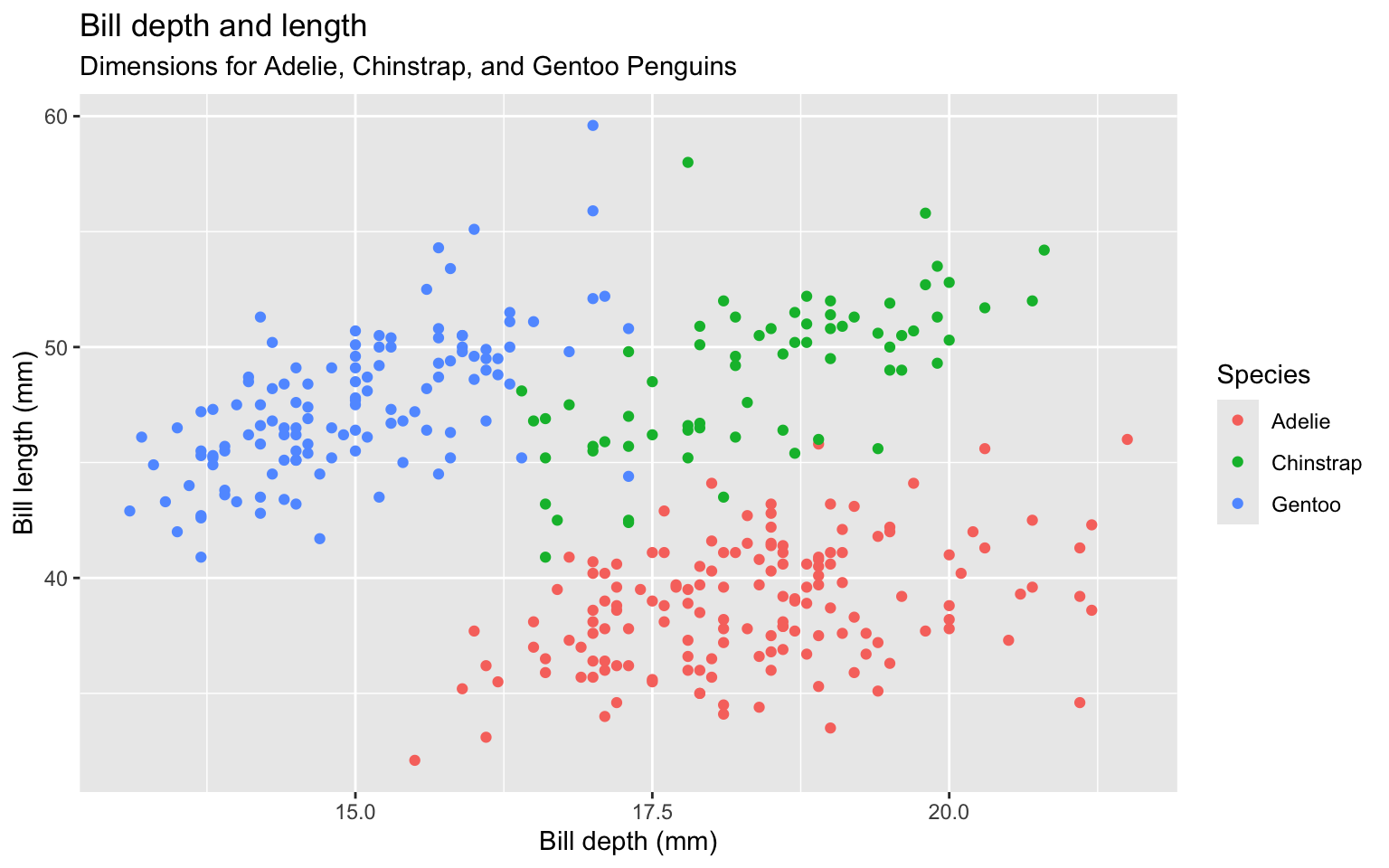
ggplot(data = penguins,
mapping = aes(x = bill_depth_mm,
y = bill_length_mm,
colour = species)) +
geom_point() +
labs(title = "Bill depth and length",
subtitle = "Dimensions for Adelie, Chinstrap, and Gentoo Penguins",
x = "Bill depth (mm)", y = "Bill length (mm)",
colour = "Species",
caption = "Source: Palmer Station LTER / palmerpenguins package") 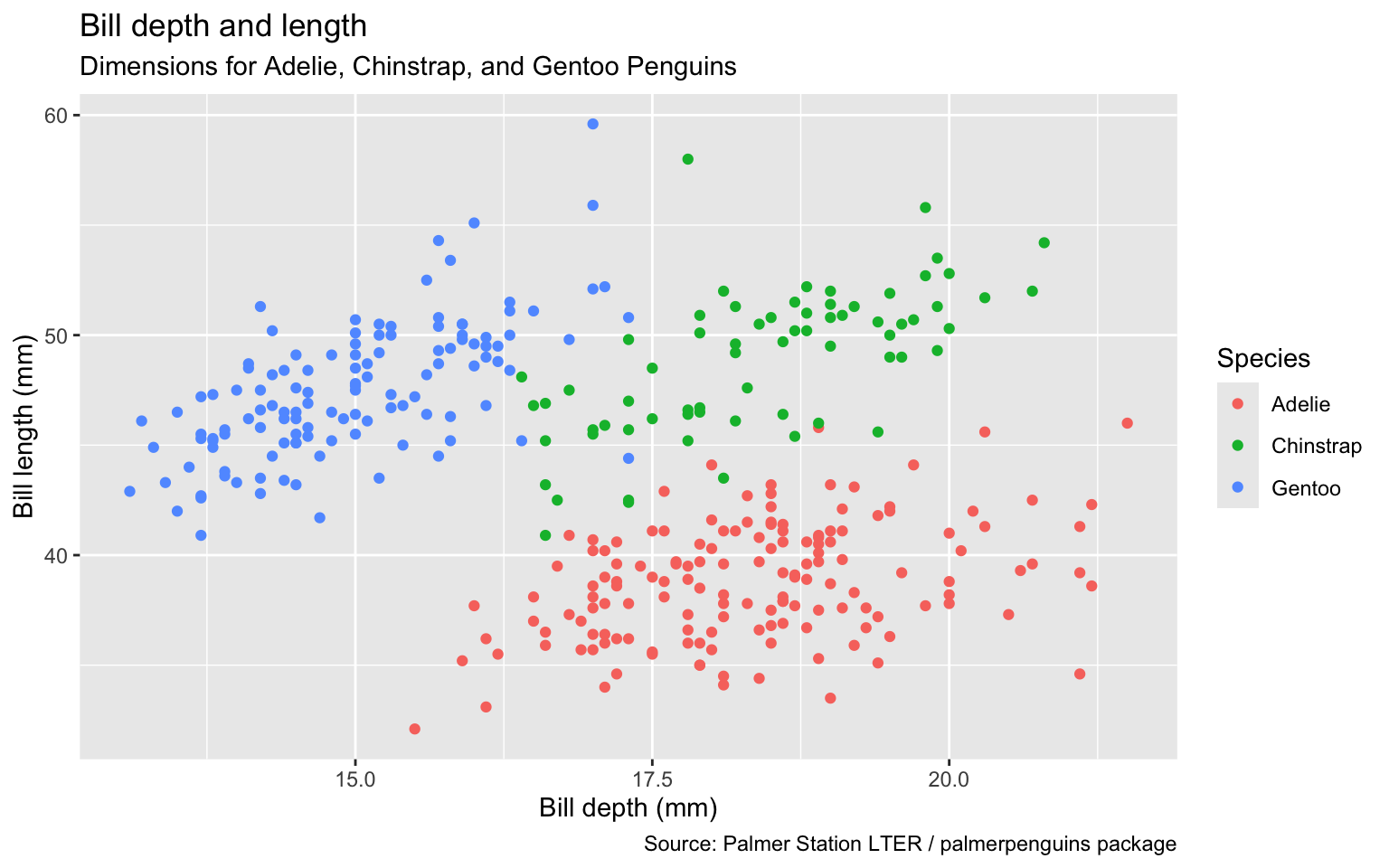
Use color scale designed to be perceived by viewers with common forms of color blindness.
ggplot(data = penguins,
mapping = aes(x = bill_depth_mm,
y = bill_length_mm,
color = species)) +
geom_point() +
labs(title = "Bill depth and length",
subtitle = "Dimensions for Adelie, Chinstrap, and Gentoo Penguins",
x = "Bill depth (mm)", y = "Bill length (mm)",
color = "Species",
caption = "Source: Palmer Station LTER / palmerpenguins package") +
scale_colour_viridis_d() 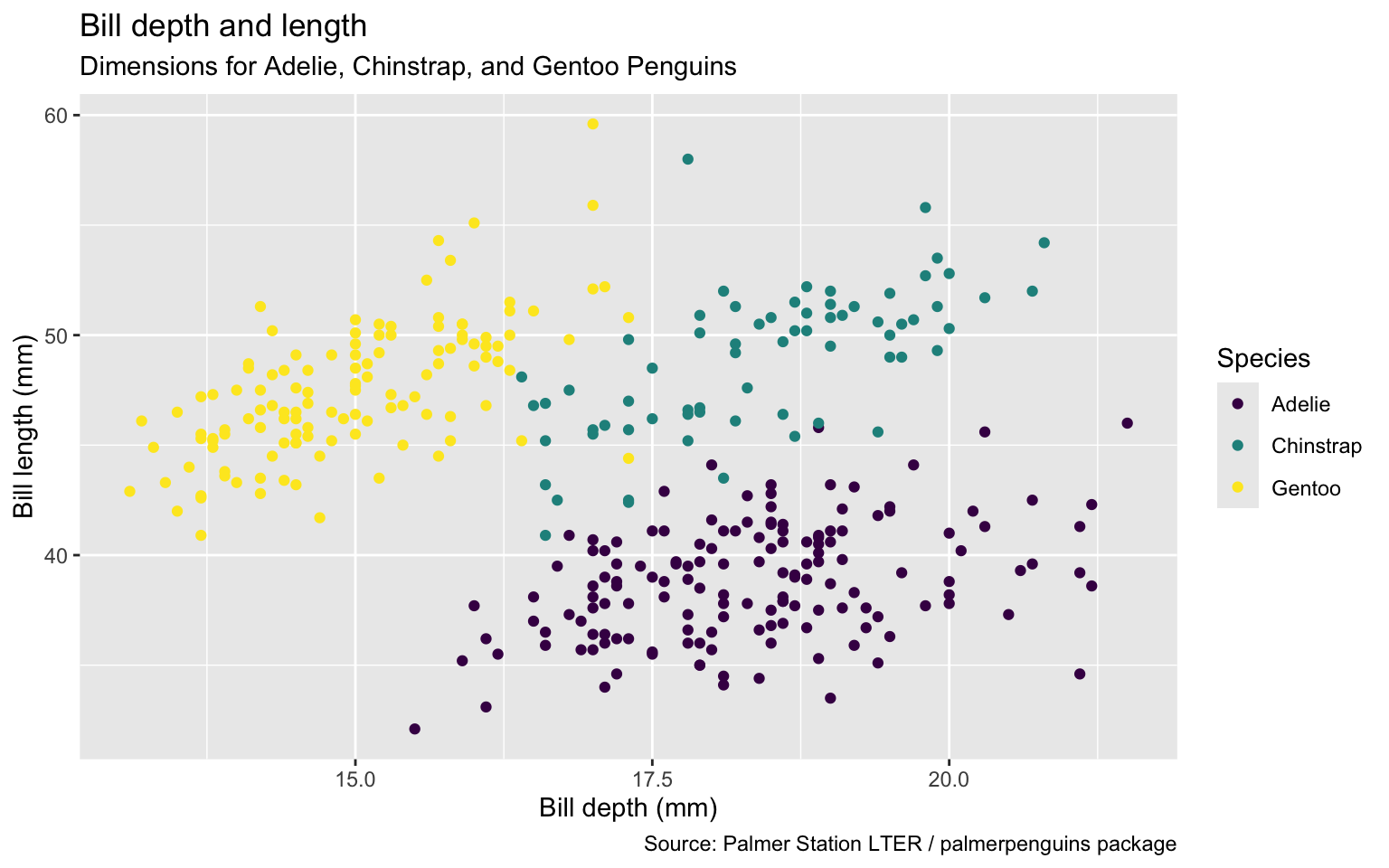
Remove argument names
ggplot(penguins,
aes(x = bill_depth_mm,
y = bill_length_mm,
color = species)) +
geom_point() +
labs(title = "Bill depth and length",
subtitle = "Dimensions for Adelie, Chinstrap, and Gentoo Penguins",
x = "Bill depth (mm)", y = "Bill length (mm)",
color = "Species",
caption = "Source: Palmer Station LTER / palmerpenguins package") +
scale_colour_viridis_d() 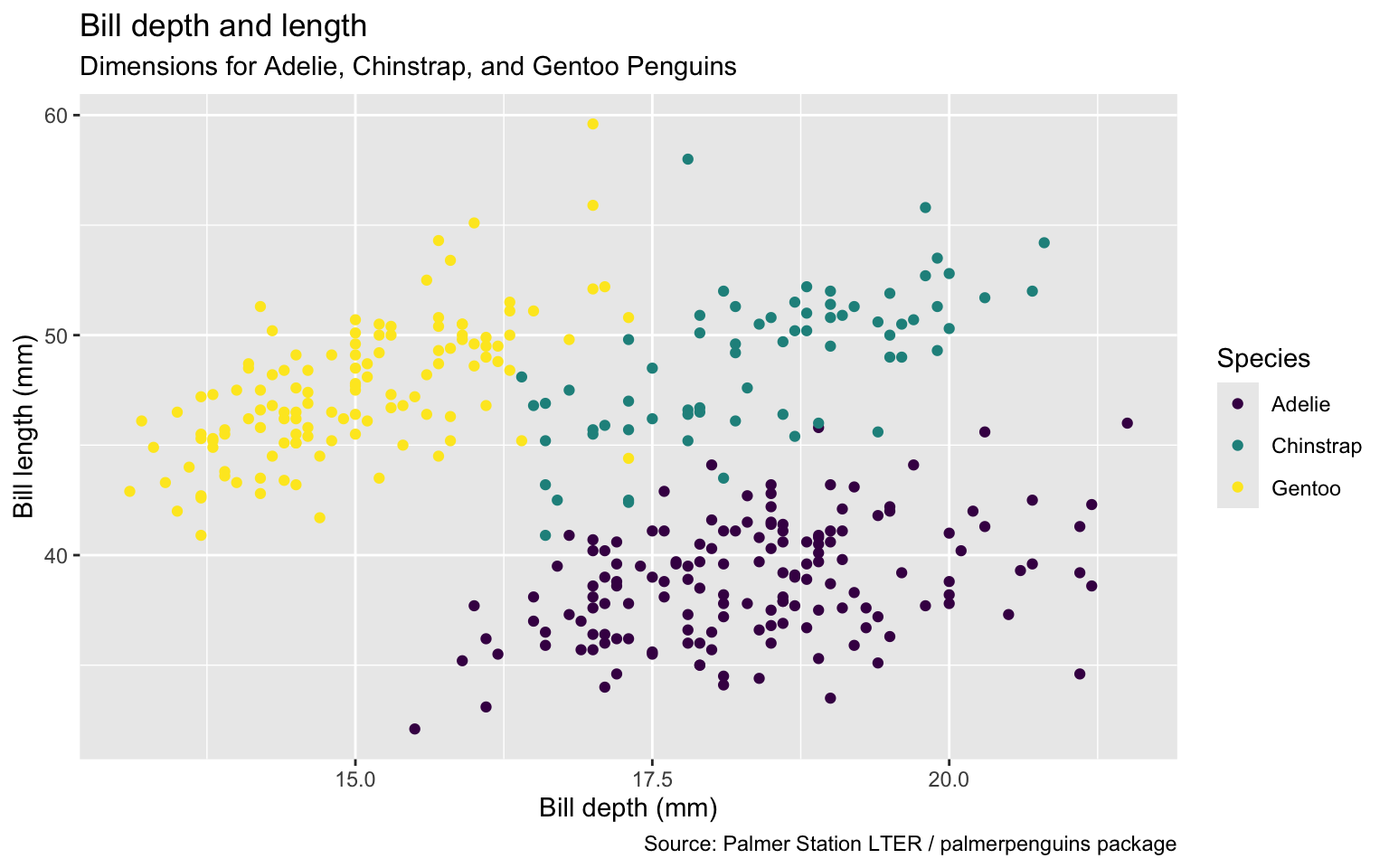
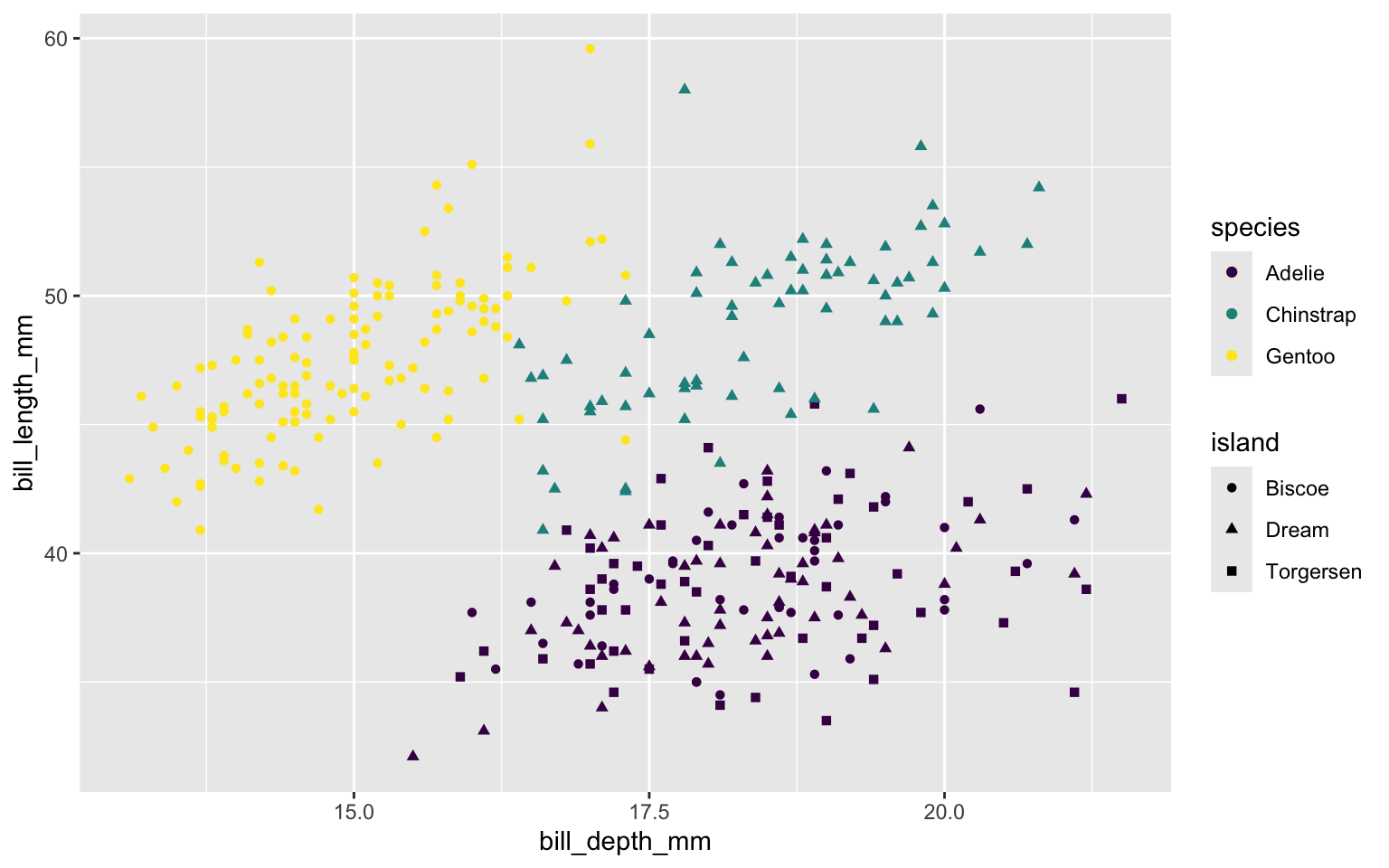
Map shapes to a different variable than we map color to. Two legends for color setting and shape setting.
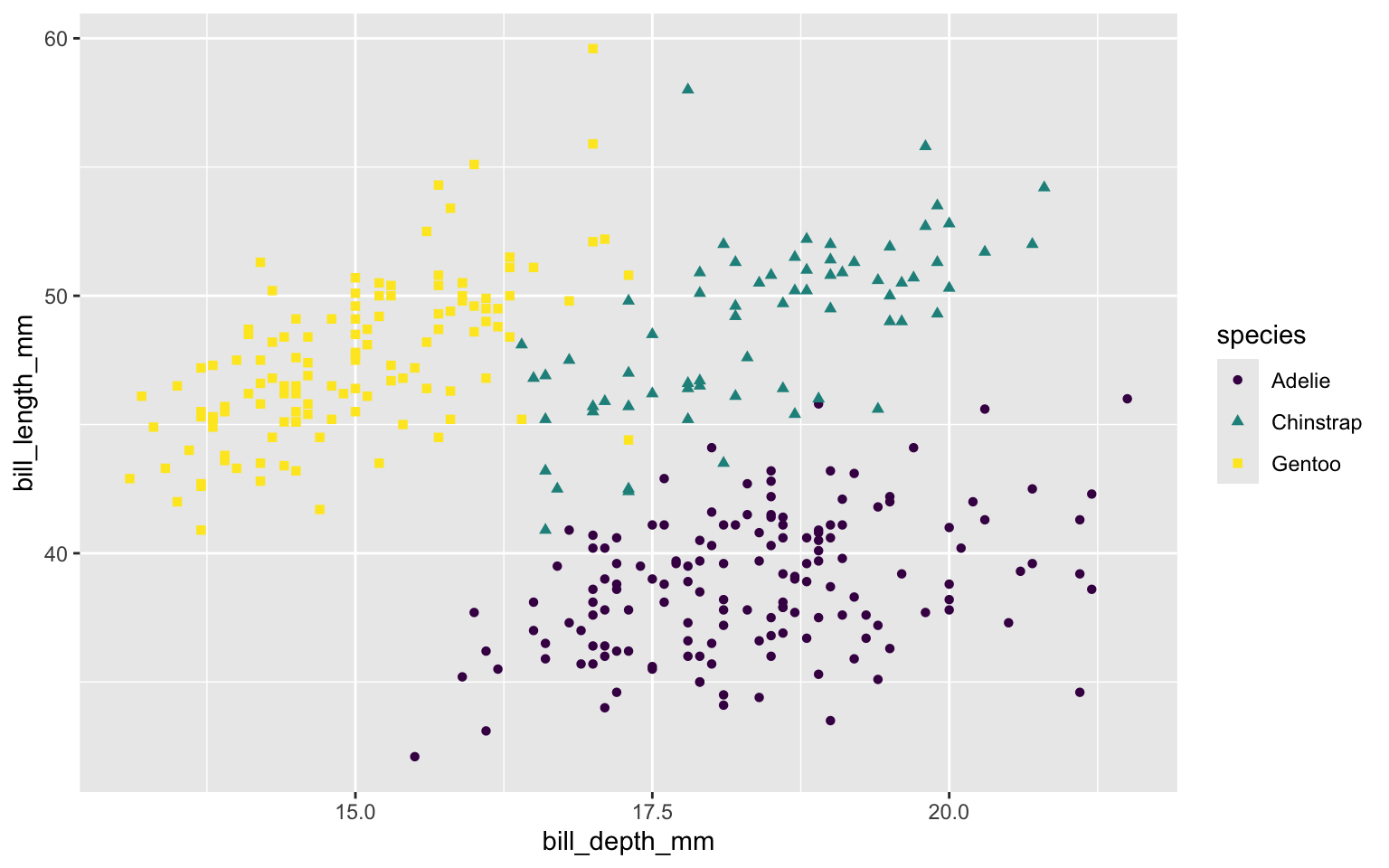
Map shape to same variable as color. Only one legend.
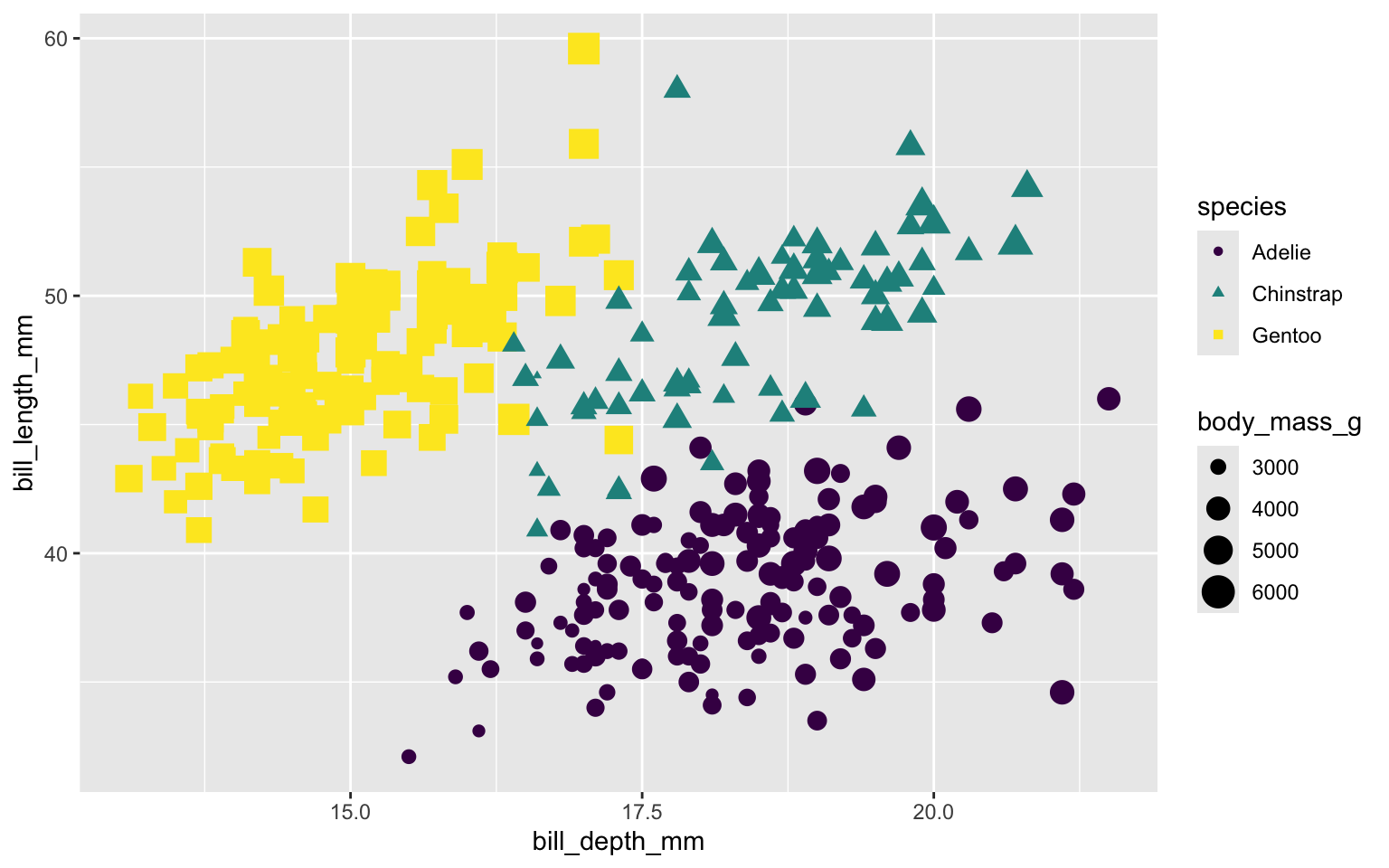
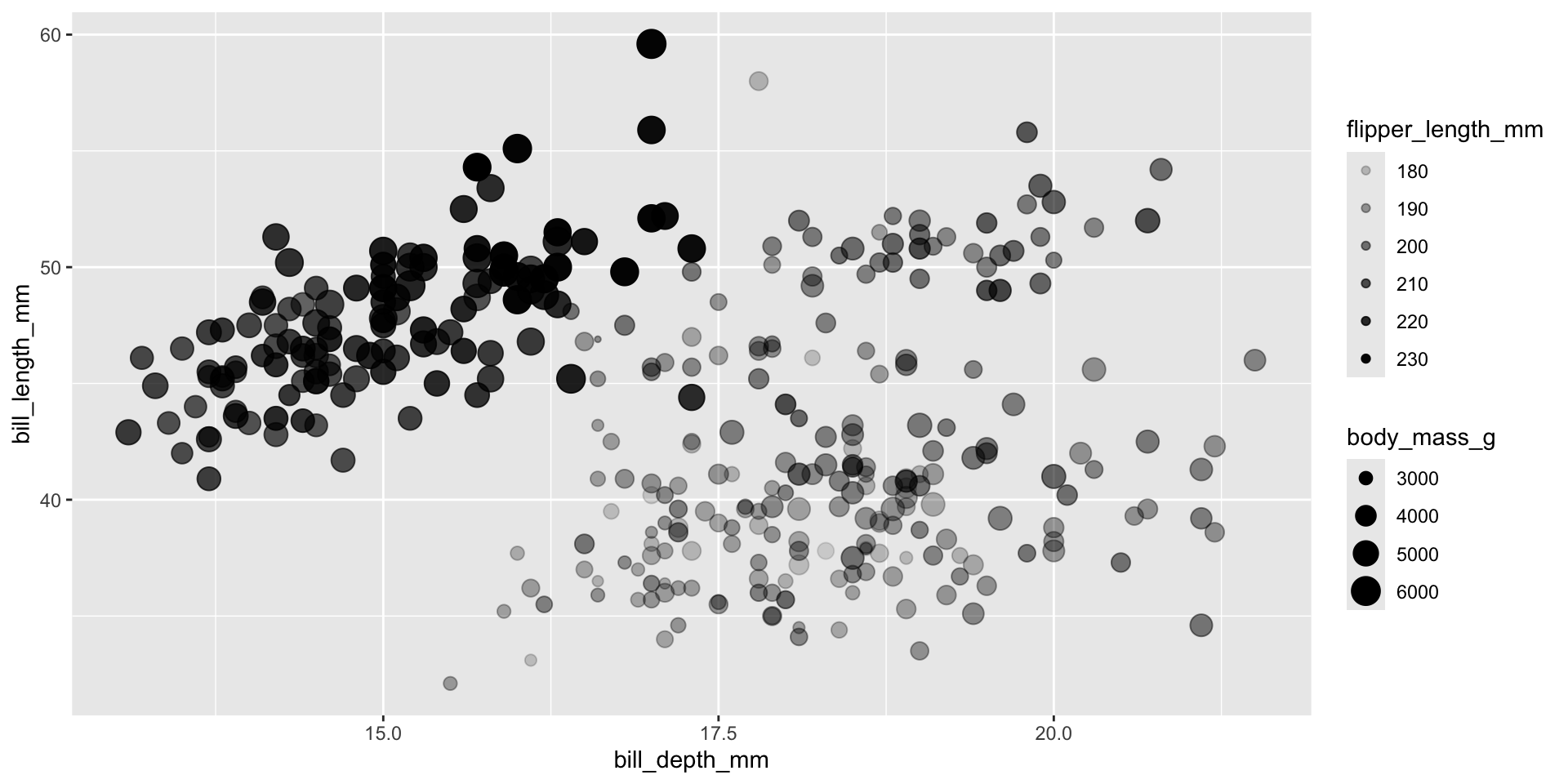
Scale size of marker based on body mass of each penguin.
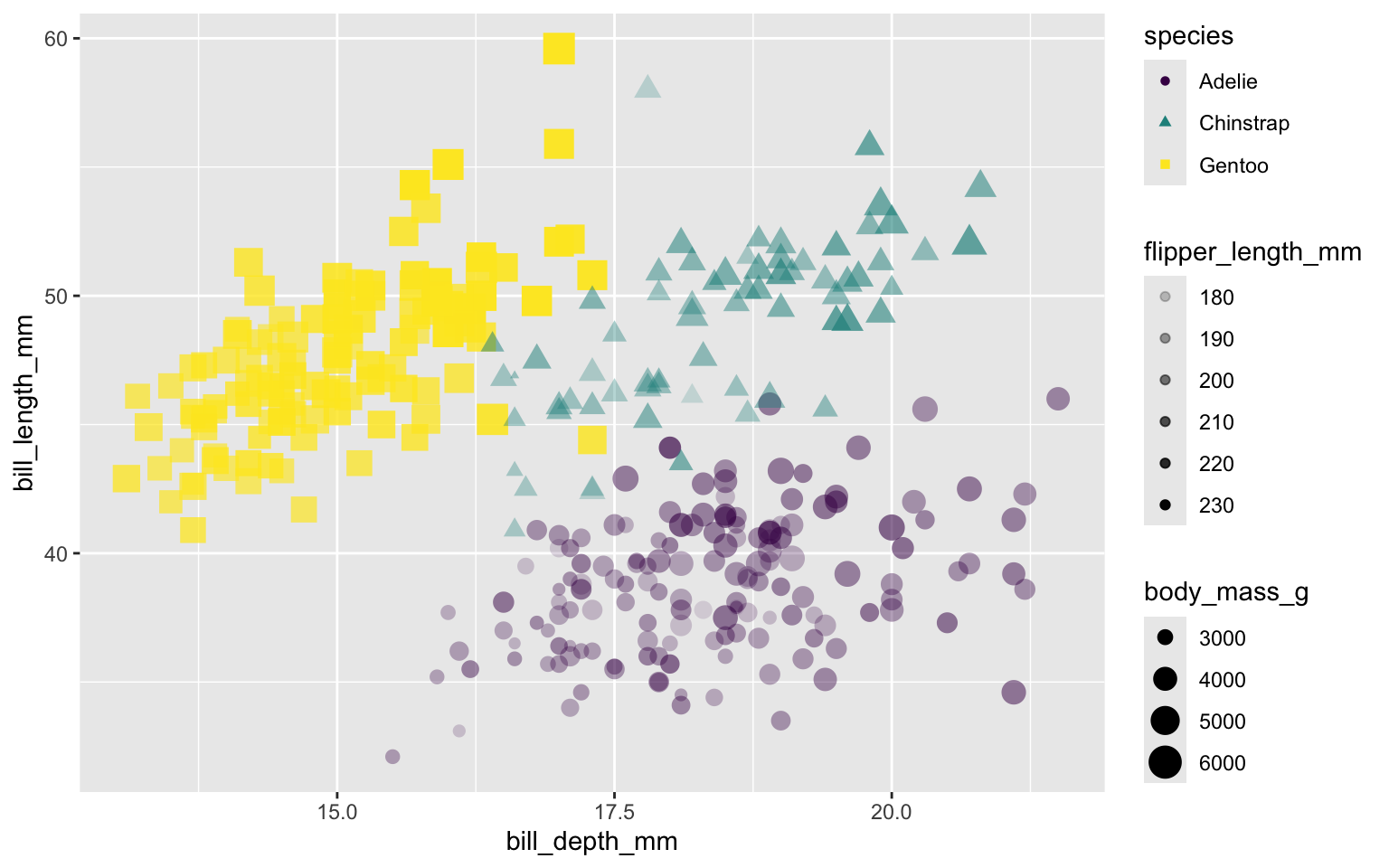
Use flipper length to control transparency of each marker – longer flipper length => darker marker
Put size and alpha in Mapping, make them dependent on two variables in the data set.
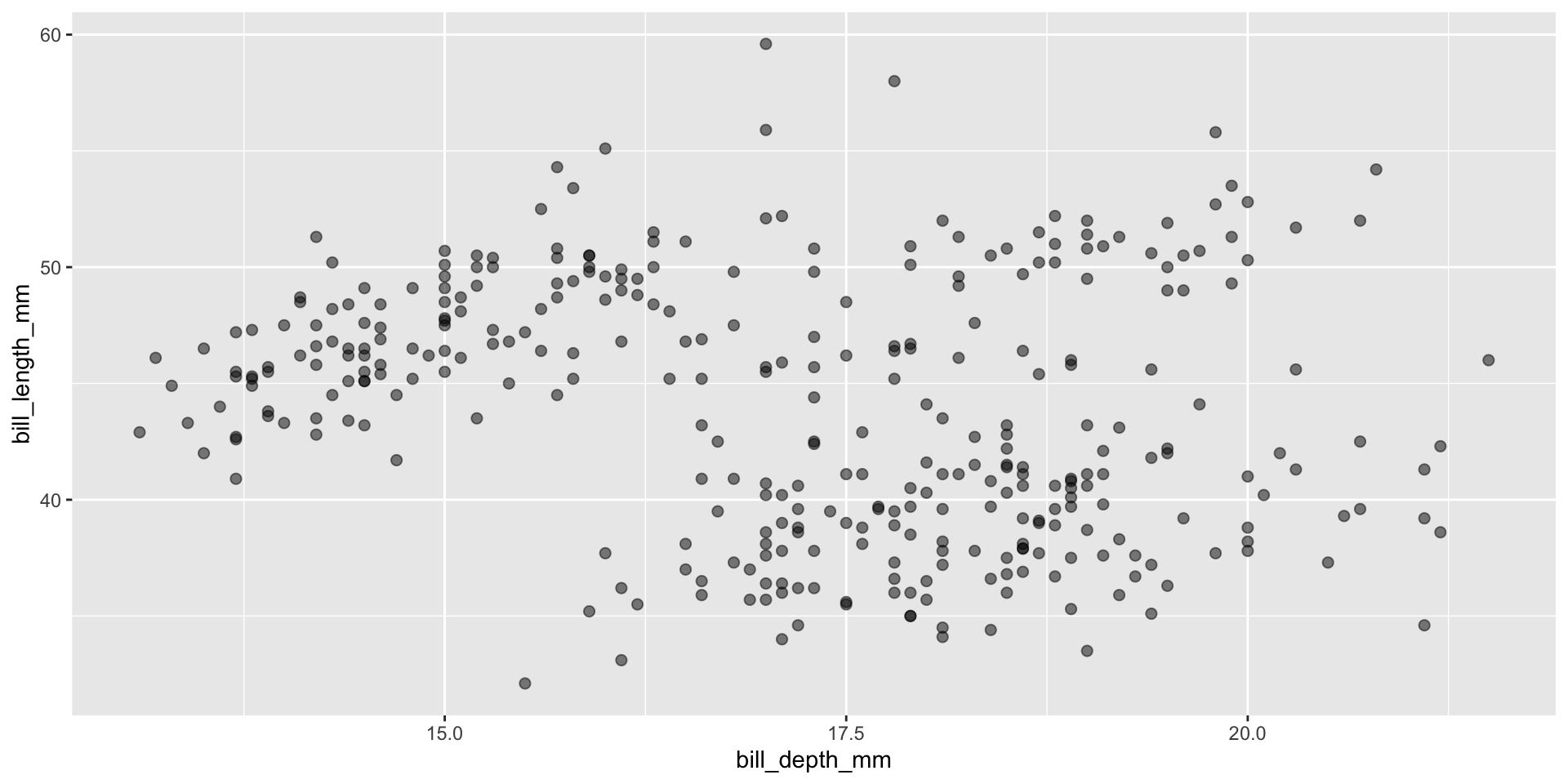
Put size and alpha into a setting instead of in the mapping
Reminder: faceting
- Small plots that display subsets of the data
- Useful for exploring conditional relationship
- Useful for exploring large datasets
- Can get complex, fast
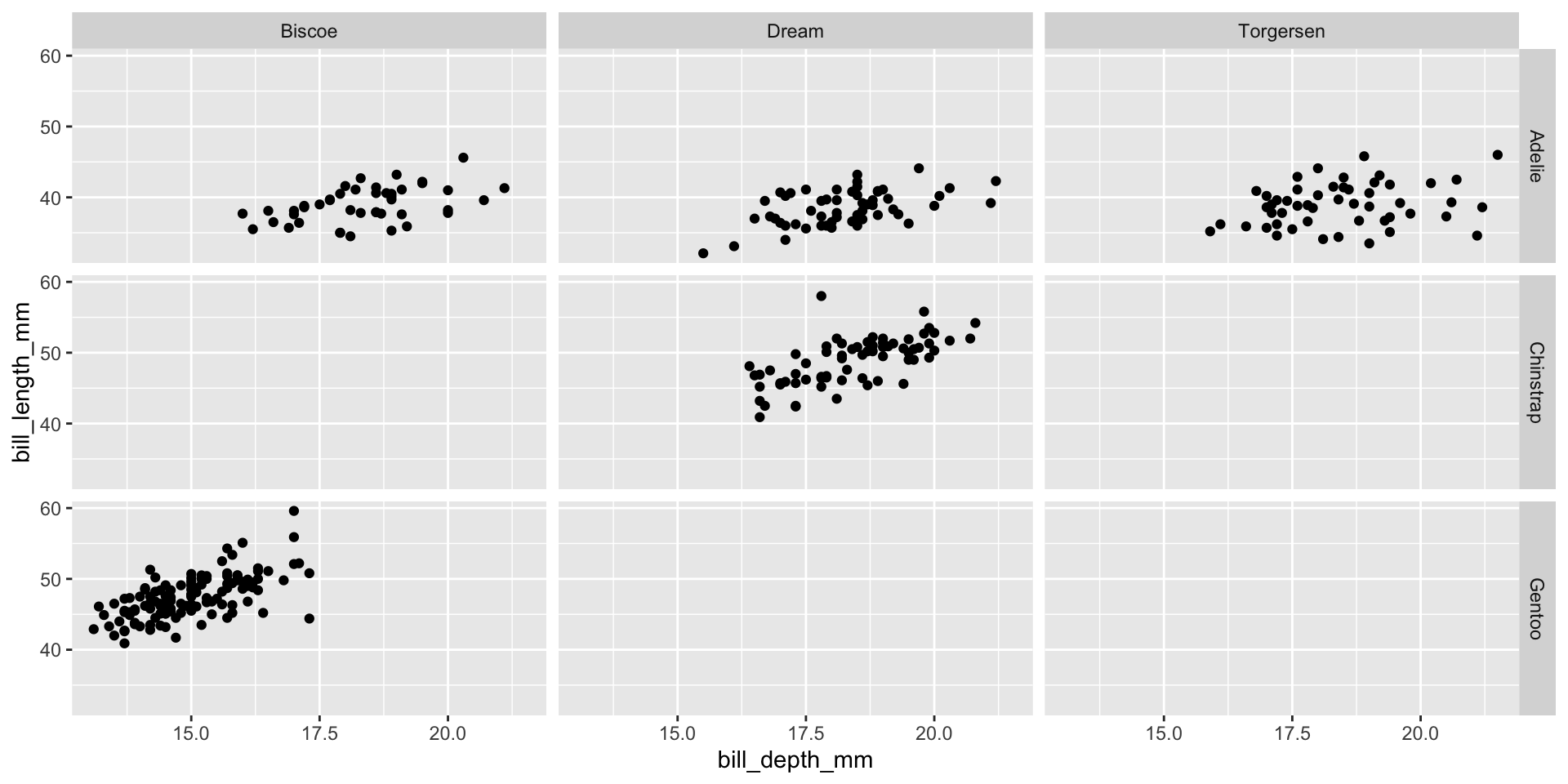
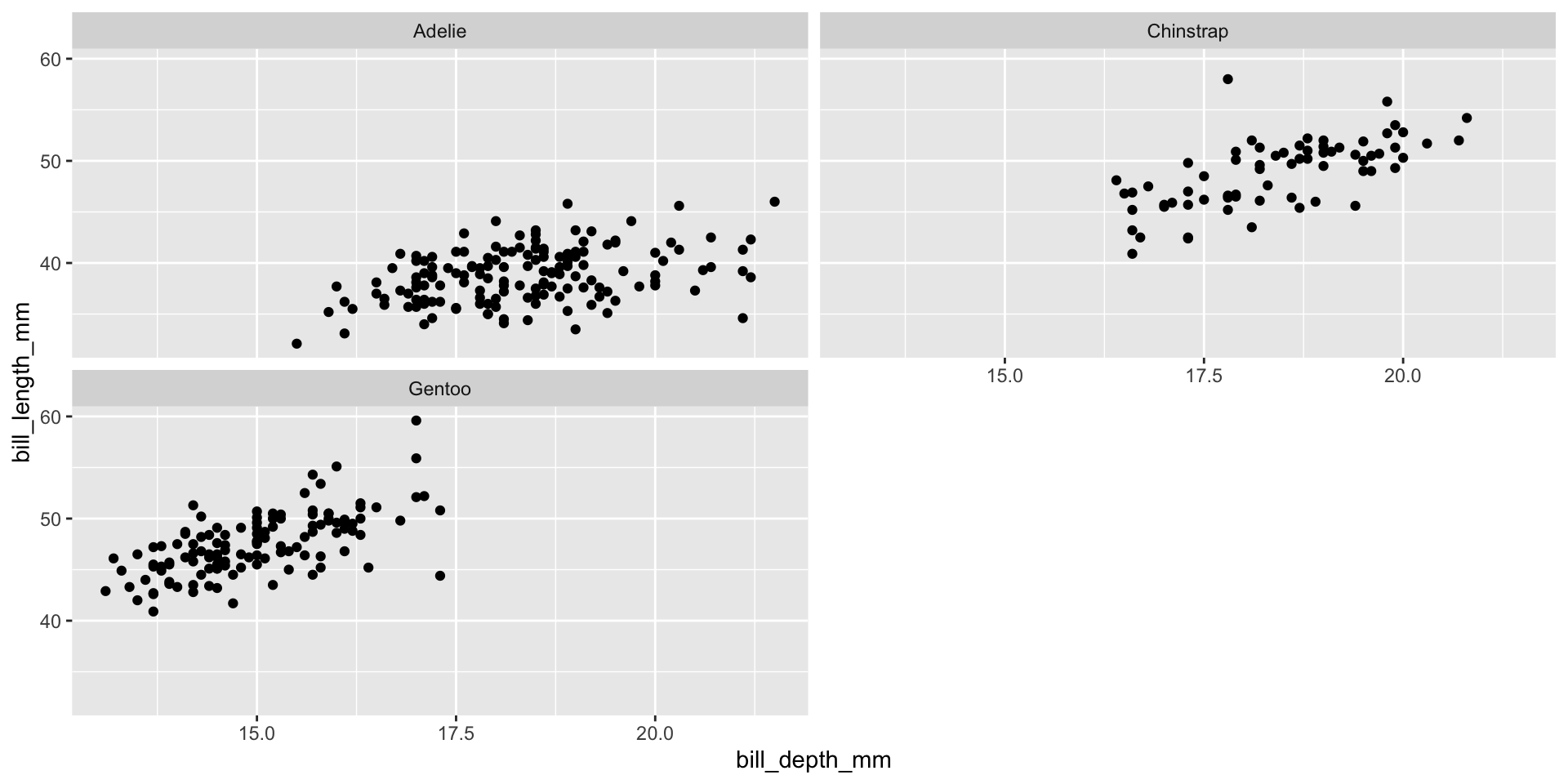
Facet example 1
Facet the plot into a grid organized by species versus island
Facet example 2
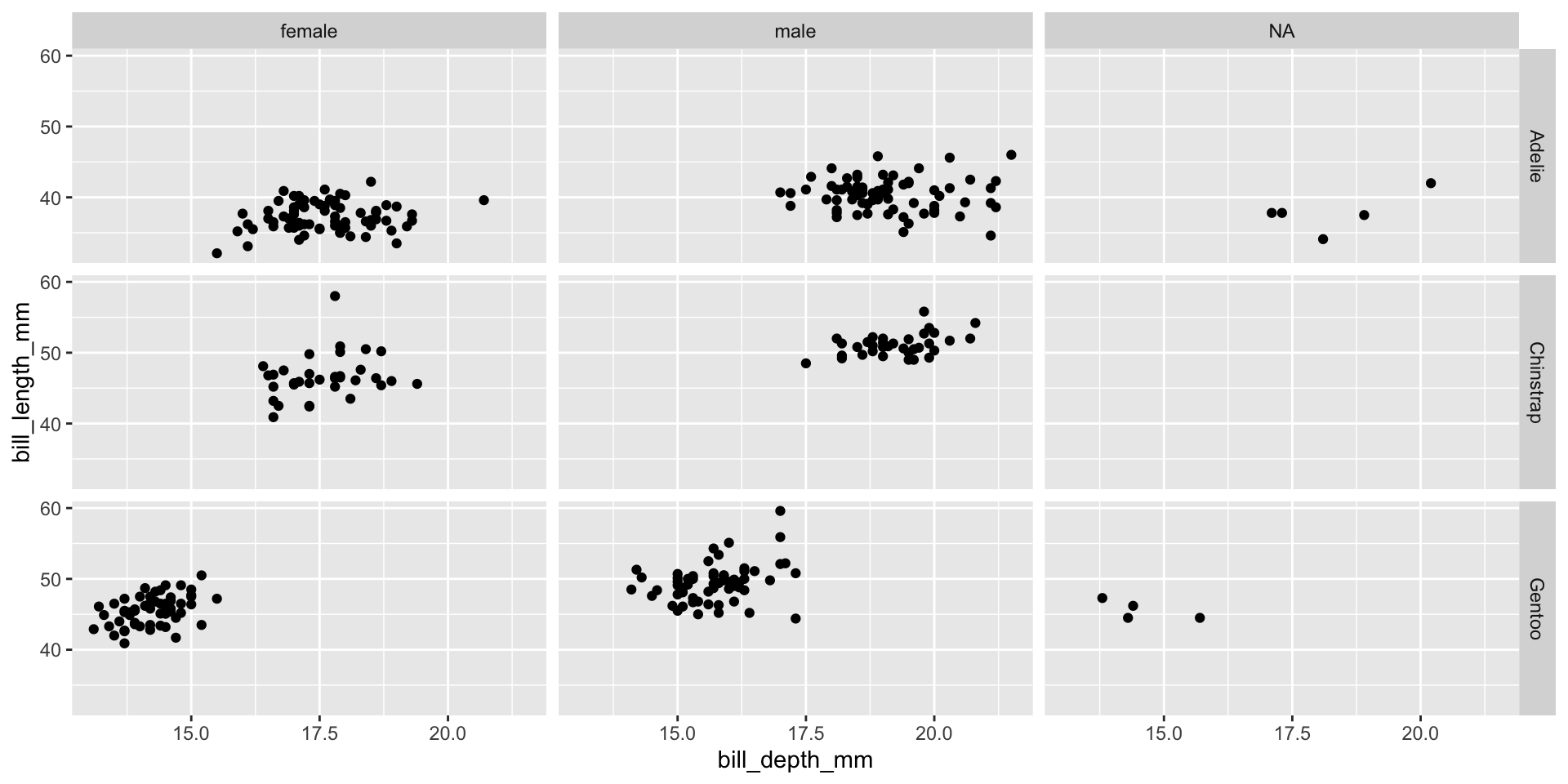
Facet example 3
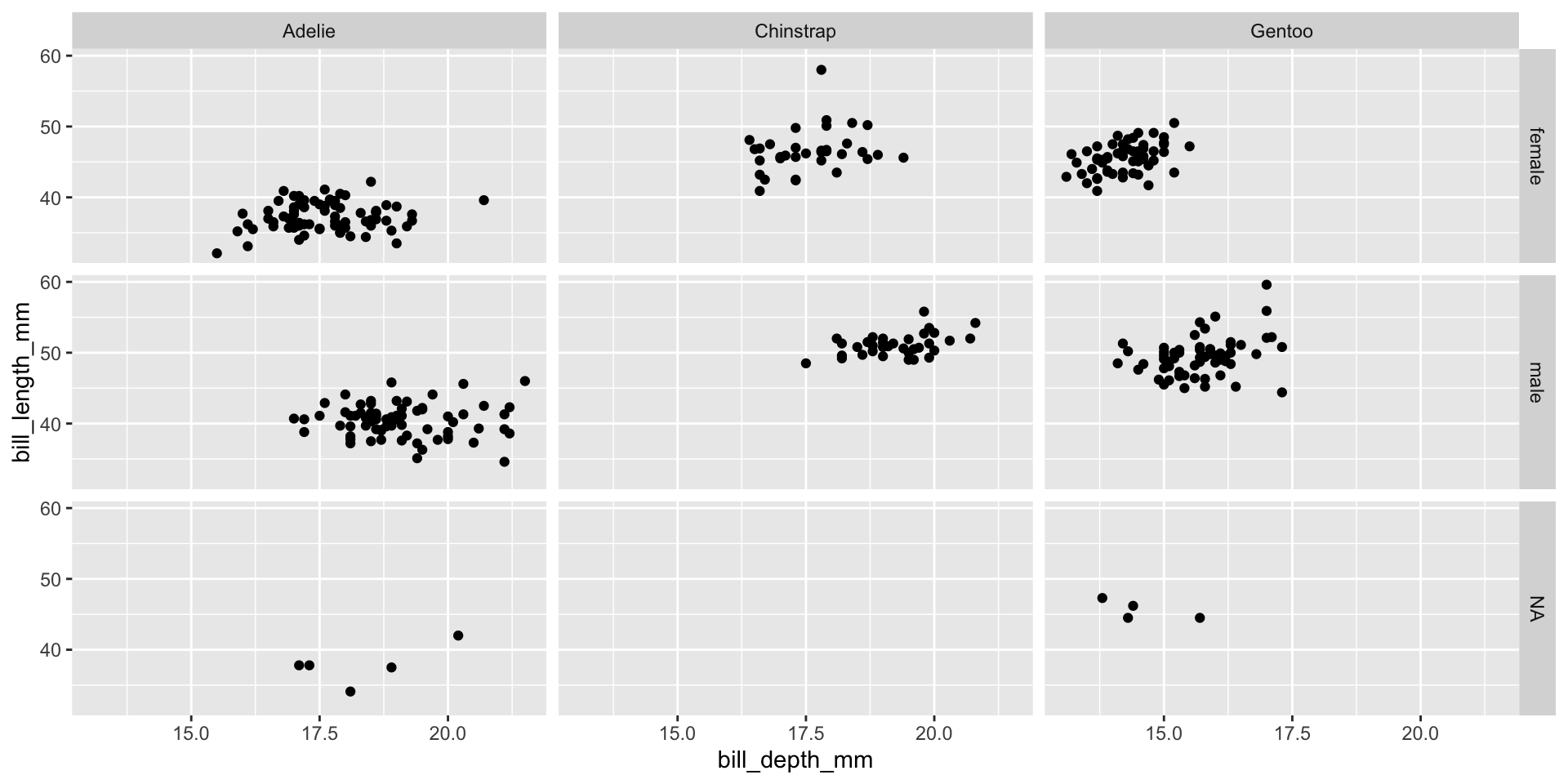
Facet example 4
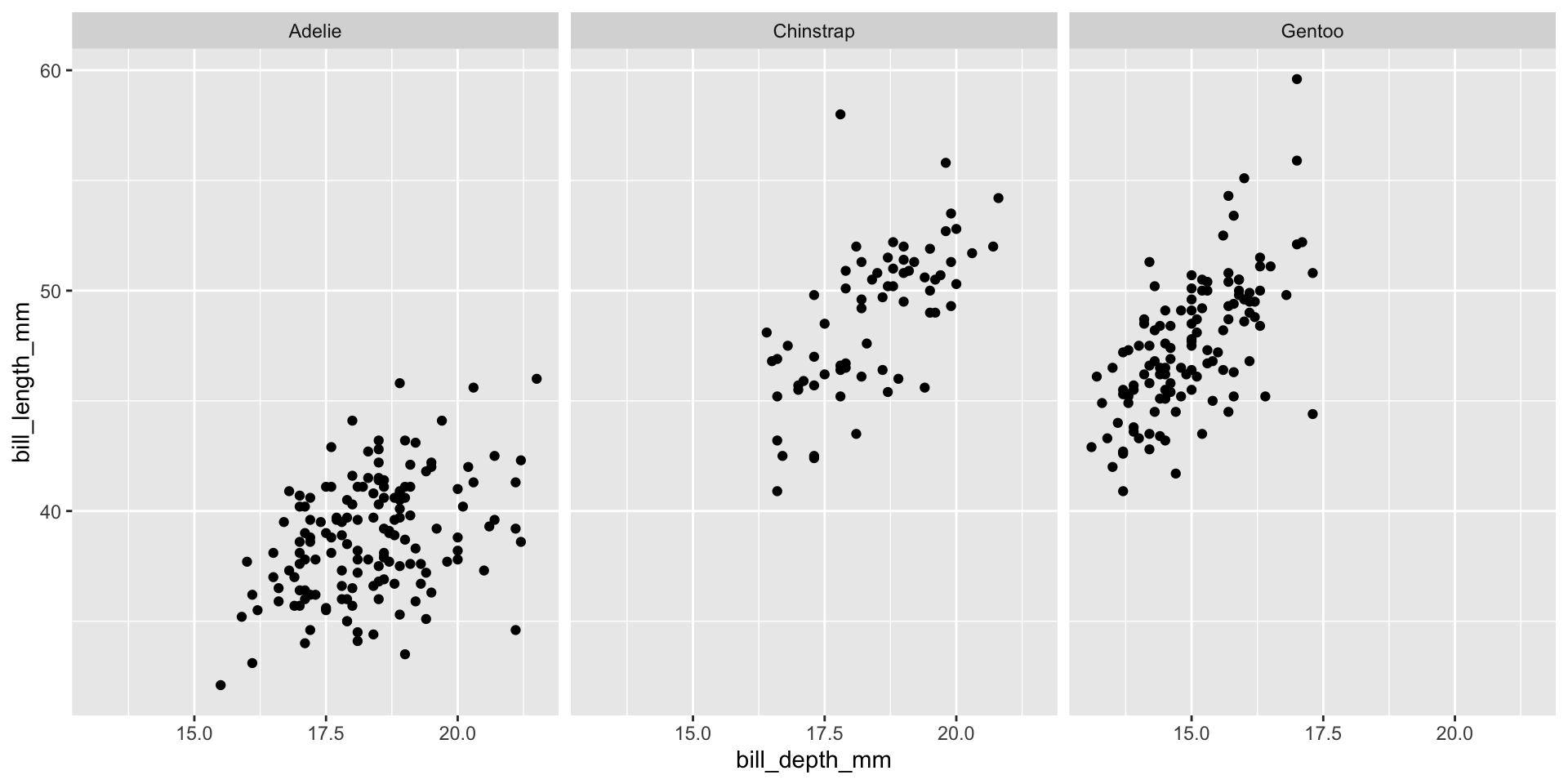
Facet example 5
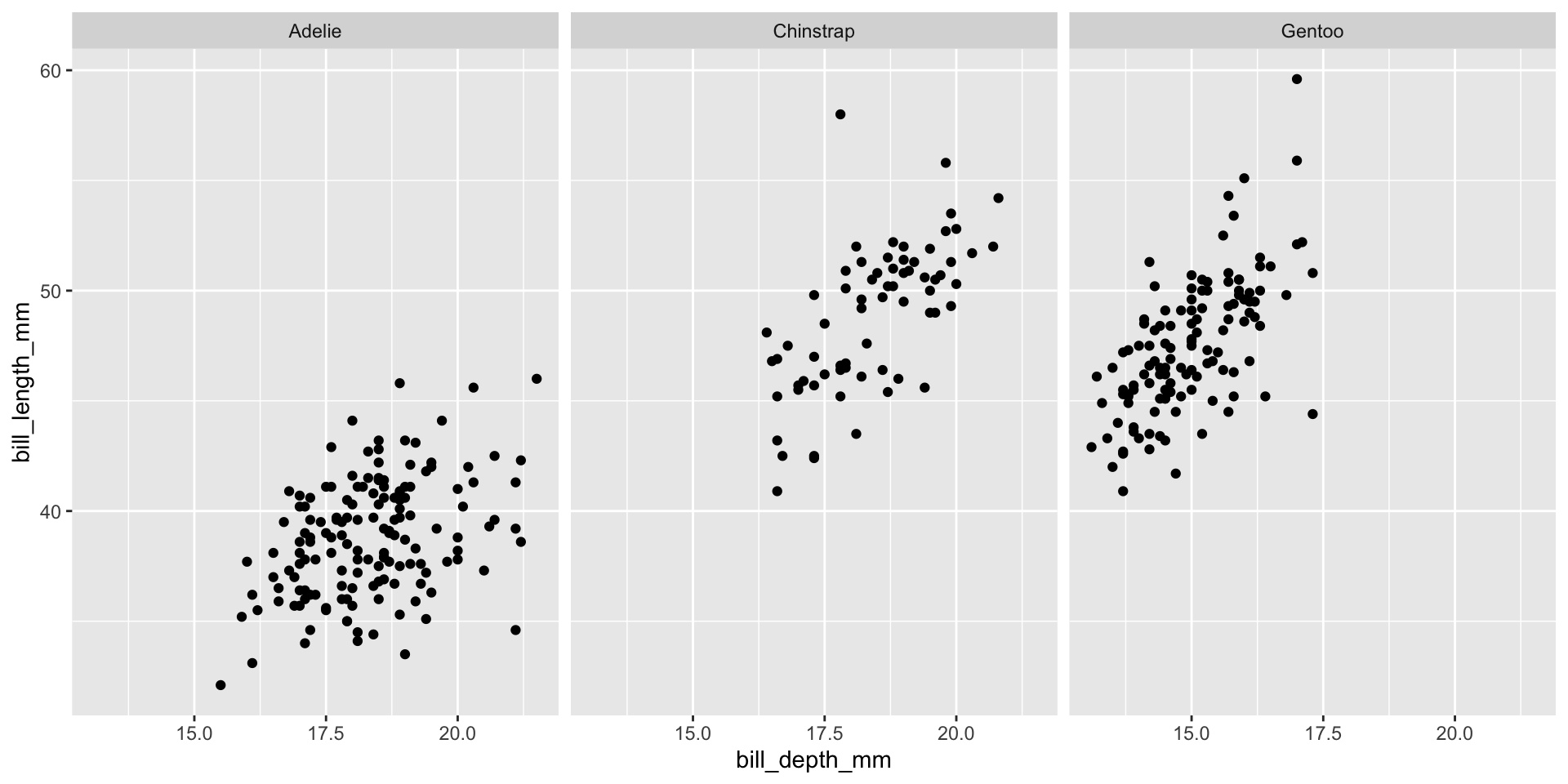
Facet example 6
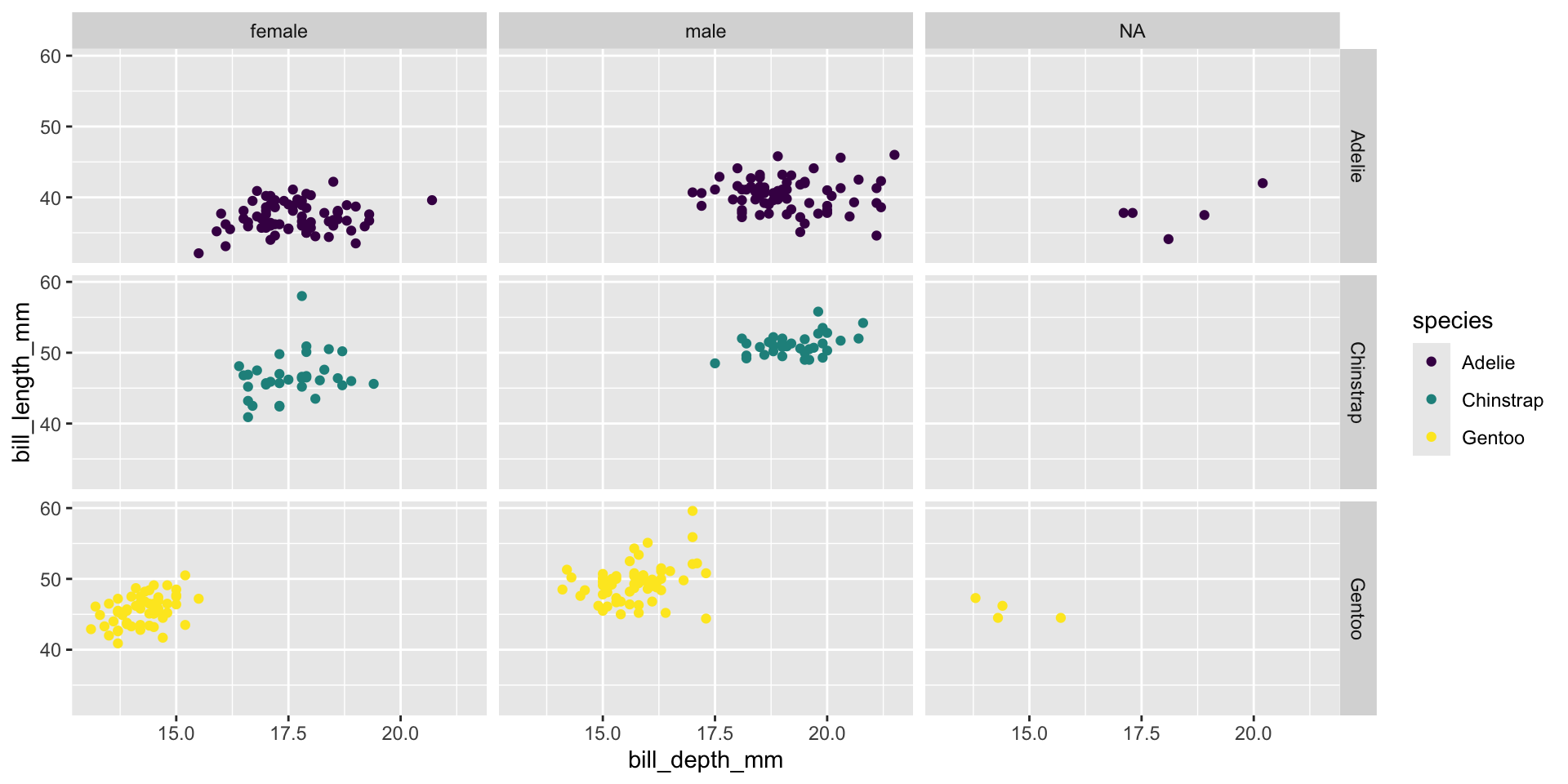
Facet and color
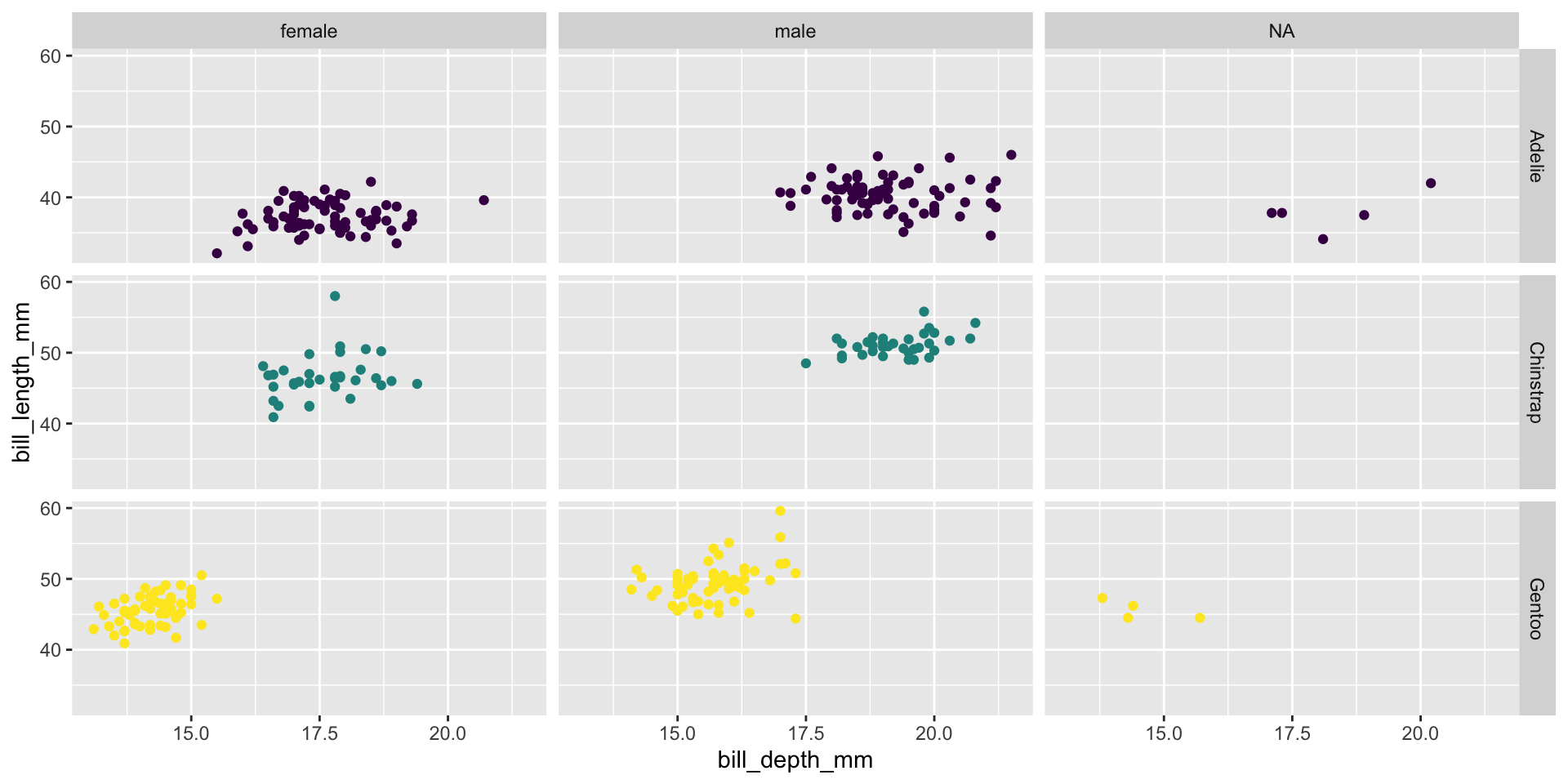
Facet and color, no legend
Faceting summary
- facet_grid():
- 2d grid
- rows ~ cols
- use . for no split
- facet_wrap():
- 1d ribbon wrapped according to number of rows and columns specified or the available plotting area